Figure 1.
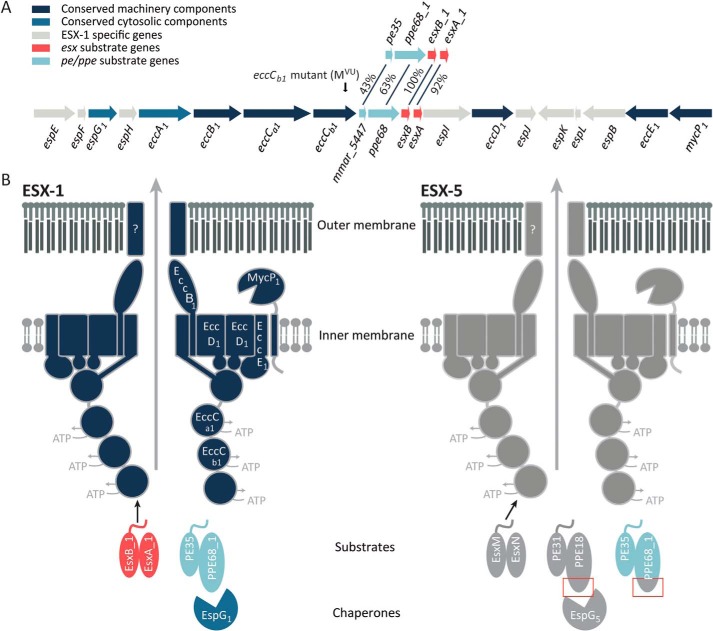
Schematic representation of the esx-1 locus and the ESX-1 and ESX-5 secretion system. A, genetic organization of the esx-1 locus and the duplicated region containing pe35/ppe68_1/esxB_1/esxA_1. Genes are color-coded according to the localization of their encoded proteins (see color key). The frameshift mutation in the eccCb1 mutant (also named MVU) used in this study is indicated by an arrow. B, current model of substrate recognition by the type VII secretion systems. Shown are the model substrates used in this study, i.e. the ESX-1 substrate pairs EsxB_1/EsxA_1 and PE35_1/PPE68_1 and the ESX-5 substrate pairs EsxM/EsxN and PE31/PPE18. Previously, PPE68_1 has been shown to be rerouted to the ESX-5 system by exchanging the EspG1-binding domain with the equivalent domain of PPE18, indicated by red boxes (32). EspG components recognize cognate PE/PPE substrates, independently of the general secretion signal located in the C-terminal tails of the PE proteins. This recognition is required for secretion via the cognate secretion machinery located in the mycobacterial inner membrane. Recognition of Esx substrates by the T7SS membrane complex occurs through the interaction of the C-terminal tail of one of the Esx partner proteins with the third nucleotide-binding domain of EccC (46, 47). How PE/PPE pairs are recognized by this complex remains unclear. Notably, only the conserved N-terminal domains of the PE and PPE proteins are shown in this model.