Figure 6.
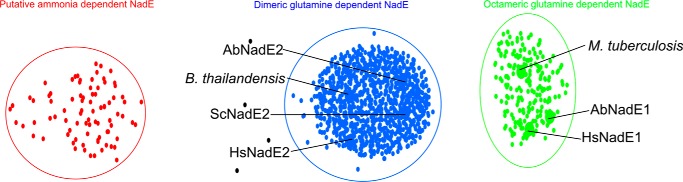
Similarity based grouping of NadE orthologues identified in the PII-NadE genomic islands. All the NadE sequences retrieved from the identified PII-NadE islands were compared using all-against-all BLAST searches using CLANS. Each protein sequence is displayed as a dot and the pairwise similarities are presented in a graph according to the similarity distances. Three major clusters were formed. The sequences of previously characterized dimeric and octameric glutamine-dependent NadE were included as function group guide in the analysis and are indicated. The green cluster included the octameric NadE1Gln from Mycobacterium tuberculosis, NadE1Gln from Herbaspirillum seropedicae (HsNadE1), and NadE1Gln from Azospirillum brasilense (AbNadE1). The blue cluster included the dimeric glutamine-dependent NadE enzymes from H. seropedicae (HsNadE2), NadE2Gln from A. brasilense (AbNadE2), NadEGln from Burkholderia thailandensis, and Synechocystis sp. (ScNadE2). The red cluster includes NadE sequences lacking the N-terminal glutaminase domain and thus were considered putative ammonia-dependent NadE.