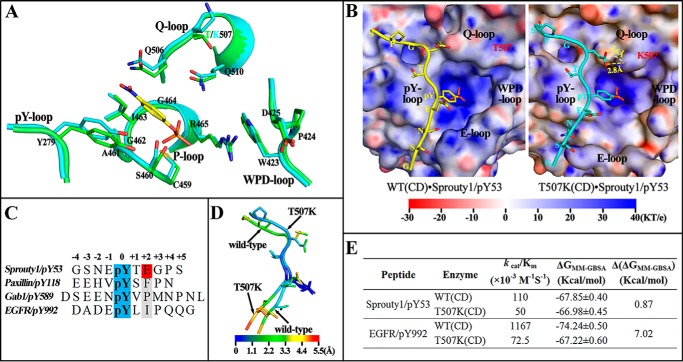
Figure 3.
Molecular basis for SHP2/T507K substrate specificity. A, comparison of active site residues in the WT SHP2 (green) and T507K mutant (cyan) with a modeled pNPP (yellow) binding at the active site. B, likely binding model for Sprouty1/pY53 peptide with SHP2 WT and T507K mutant catalytic domain. The electrostatic potential was mapped on the protein surface, and residues Thr-507 and Lys-507 are shown as sticks below the slightly transparent surface. C, sequence comparison among four pTyr-containing peptide substrates. D, atom fluctuation heat map for Sprouty1/pY53 peptide during a 5-ns MD simulation of two models in B. E, binding free energies for four indicated systems were calculated by MM-GBSA method.