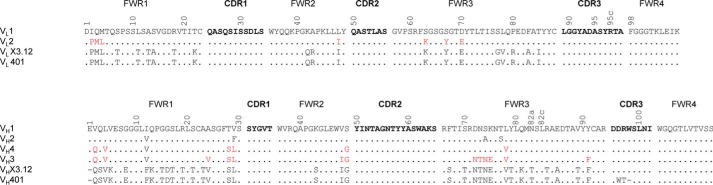
Figure 2.
Amino acid sequence alignment of VL and VH of the affinity-matured and humanized variants. The location of the four framework regions (FWRs) and the three CDRs is indicated. The numbers refer to Kabat numbering of variable domain residues shown in single-letter code. Residues shown in red were back-mutated to the original rabbit residue. Dots, identical residues in the alignment to VL1 or VH1. CDR residues are shown in boldface type. Top, the VL1 FWRs are derived from human germline IGKV1-NL1*01; the VL1 CDRs are grafted from X3.12. The VL2 amino acid sequence is the same as VL1 but with seven back-mutations from the human germline to the original rabbit residues of X3.12. Bottom, the VH1 and VH2 FWRs are derived from human germlines IGHV3–66*03 and IGHV3–48*03, respectively, and their CDRs are grafted from X3.12. VH3 and VH4 vary from VH1 and VH2 by back-mutating FWR residues from the human germline to the original rabbit residues of X3.12.