Figure 1.
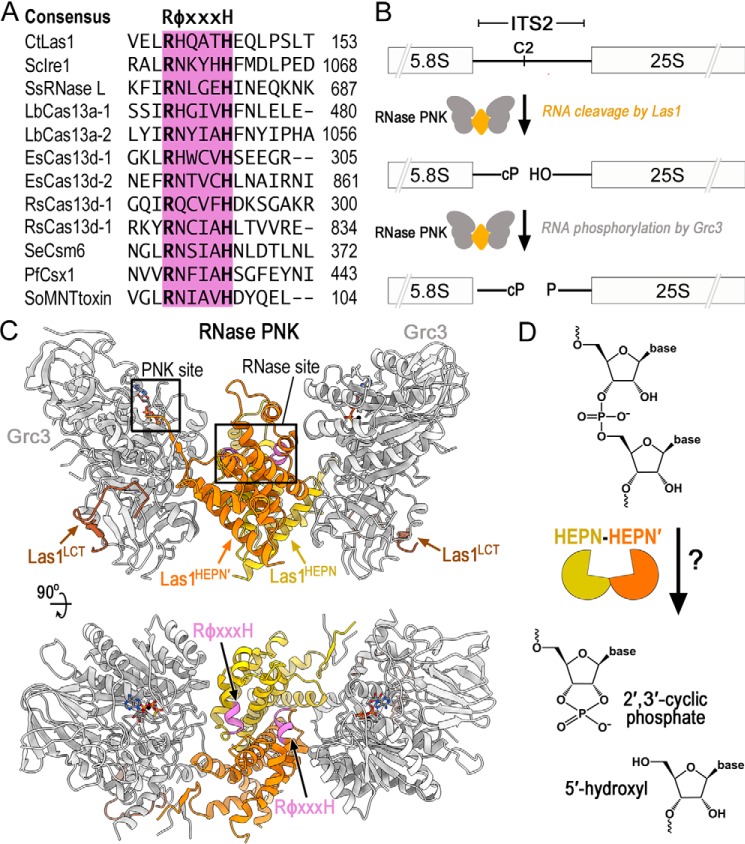
Members of the HEPN superfamily encode a canonical RφXXXH motif responsible for RNA cleavage. A, amino acid sequence alignment of the HEPN motif from several different HEPN nucleases. The HEPN canonical motif RφXXXH (where φ is commonly N, H, or D and X is any residue) is responsible for RNA cleavage and highlighted in purple. Abbreviations are as follows: C. thermophilum (Ct), S. cerevisiae (Sc), Sus Scrofa (Ss), Leptotrichia buccalis (Lb), Eubacterium siraeum (Es), Ruminococcus species (Rs), Staphylococcus epidermidis (Se), Pyrococcus furiosus (Pf), and Shewanella oneidensis (So). B, diagram of ITS2 pre-rRNA processing by RNase PNK. RNase PNK is composed of the Las1 RNase and the Grc3 PNK. Las1 cleaves the C2 site leaving a 2′,3′-cyclic phosphate (cP) and a 5′-hydroxyl (OH) (14). Subsequently, Grc3 phosphorylates (P) the 5′-hydroxyl end marking the ITS2 for decay by a series of exoribonucleases. C, orthogonal views of C. thermophilum RNase PNK model (PDB ID 6OF3). Grc3 promoters are shown in gray and Las1 protomers are shown in orange and yellow. Las1 RφXXXH motifs are highlighted in purple and ATP-γS are shown as sticks. Las1 RNase and Grc3 PNK active sites are boxed. D, cartoon schematic of metal-independent RNA cleavage by members of the HEPN superfamily. HEPN members dimerize (orange/yellow) to form an active nuclease, which cleaves the phosphodiester backbone through an unclear mechanism resulting in a 2′,3′-cyclic phosphate and 5′-hydroxyl RNA ends.