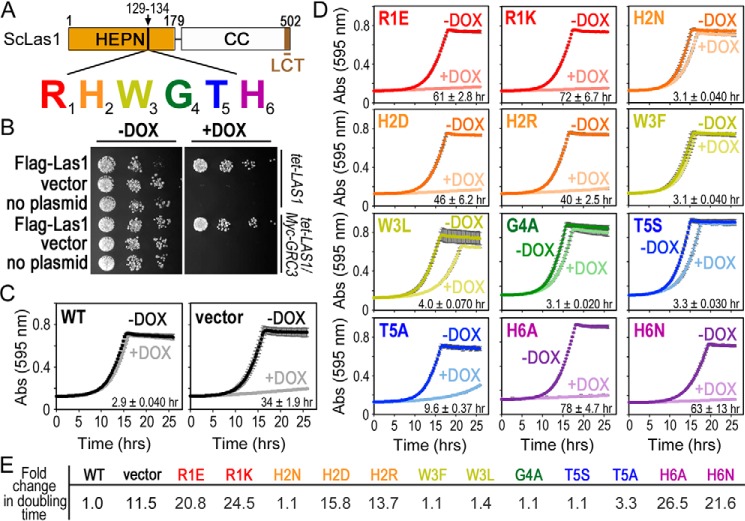
Figure 3.
The Las1 RHXhTH motif is essential for S. cerevisiae proliferation. A, cartoon of S. cerevisiae (Sc) Las1 with the numbers defining the amino acid residue domain boundaries. The Las1 HEPN domain is shown as an orange box, the coiled-coil (CC) domain is a white box and the Las1 C-terminal tail (LCT: residues 469–502) is shown in brown. Individual residues of the ScLas1 RHXhTH motif (129RHWGTH) are highlighted in red, orange, yellow, green, blue, and purple, respectively. B, S. cerevisiae tet-LAS1 and tet-LAS1/Myc-GRC3 strains were either left untransformed (no plasmid) or transformed with plasmids encoding WT Las1 with an N-terminal 3× FLAG tag (FLAG-Las1) and empty YCplac 33 vector (vector). Serial dilutions of each strain were grown on YPD agar in the absence (−DOX) and presence (+DOX) of 40 μg/ml doxycycline at 30 °C for 3 days. Doxycycline represses transcription of endogenous LAS1 (Fig. S2A). C, growth curves of tet-LAS1/Myc-GRC3 strains transformed with control plasmids encoding WT FLAG-Las1 (WT) and empty YCplac 33 (vector). Strains were incubated at 30 °C in the absence and presence of doxycycline and absorbance was recorded over 25 h at 595 nm. Each curve is an average of three independent replicates and error bars define the S.D. D, growth curves as seen in panel C with the tet-LAS1/Myc-GRC3 strain transformed with FLAG-Las1 RHXhTH variants (Table S2). Color scheme is the same as seen in panel A. The doubling time was calculated using the +DOX curve and shown in the lower right corner. The average doubling time and S.D. were calculated from three biological replicates. E, -fold change in the doubling time for the different yeast strains normalized to the WT strain.