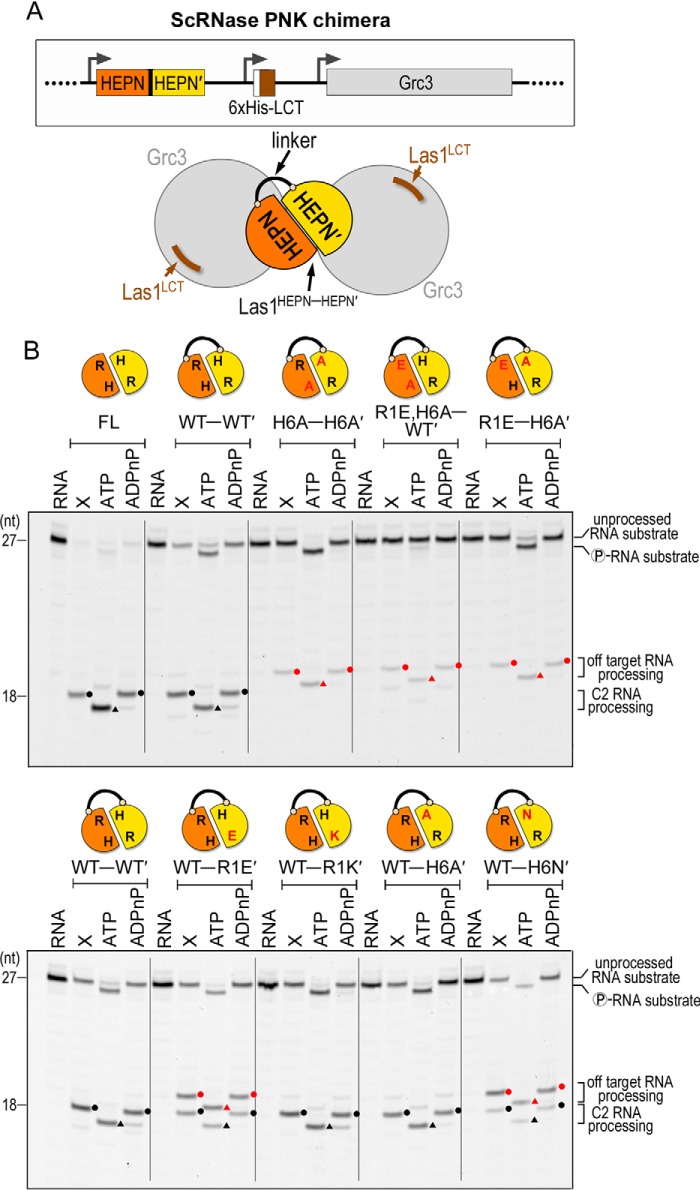
Figure 5.
Las1 HEPN-HEPN′ chimeras have altered cleavage fidelity. A, cartoon schematic of the chimeric ScRNase PNK complex comprised of the Las1 HEPN-HEPN′ dimer (orange-yellow) connected by a flexible linker, the Las1 LCT (brown) and WT Grc3 (gray). The black line represents the linker tethering the Las1 HEPN-HEPN′ dimer. The Las1 coiled-coil domain is dispensable for C2 RNA processing in vitro (23) and was removed to create the chimeric ScLas1HEPN-HEPN′+LCT-Grc3 complex. B, denaturing urea gels of RNA cleavage and phosphorylation reactions with full-length RNase PNK (FL) and chimeric RNase PNK HEPN variants (HEPN-HEPN′). The lanes are marked as follows: RNA (100 nm RNA alone), X (0.8 μm RNase PNK variant and 100 nm RNA), ATP (0.8 μm RNase PNK variant, 100 nm RNA, and 10 mm ATP), ADPnP (0.8 μm RNase PNK variant, 100 nm RNA, and 10 mm ADPnP). Circles mark RNA cleavage products and triangles identify phosphorylated RNA. Black symbols specify C2 RNA processing products and red symbols are off-target RNA processing products.