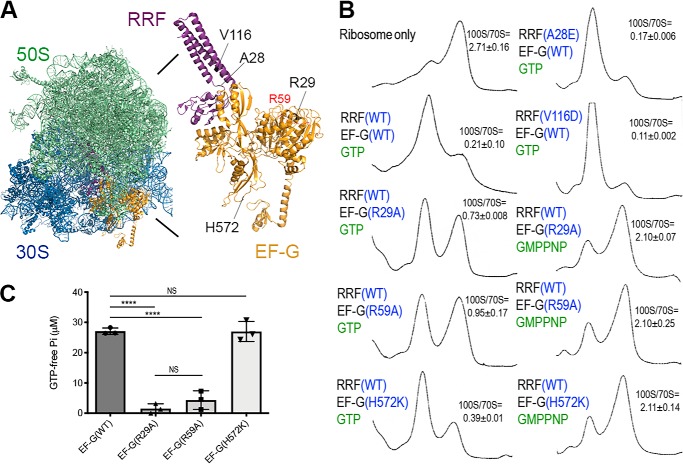
Figure 2.
The effects of RRF and EF-G mutations in 100S ribosome disassembly. A, model of the RRF, EF-G and ribosome co-complex. A structural model was constructed from PDB accession numbers 4V54, 5OT7, and 4WPO. The mutations included in this study are indicated. R59 (red) was mapped to an unstructured region of EF-G in the crystal. B, the dissociation of 100S ribosomes is reduced in GTP-hydrolysis mutants (R29A and R59A) of EF-G but is unaffected by a mutation that compromises tRNA translocation (H572K). The dissociation reactions comprised 0.2 μm ribosomes, 2 μm proteins, and 2 mm GTP analogs and were incubated at 37 °C for 30 min. Ribosome profiles were analyzed via 5–20% sucrose density sedimentation, and the ribosomal species were monitored according to the absorbance at 254 nm. 100S to 70S ratios were obtained from three technical replicates (of two independently prepared ribosomes and recombinant proteins); mean ± S.D. C, malachite green GTPase assay showing the reduction of GTPase hydrolysis in the R29A and R59A mutants. The known translocation inactive H572K mutant does not present impaired GTPase activity. Error bars represent the S.E. obtained from three independent experiments using two different batches of purified proteins. p values were calculated by Student's unpaired t test, ****, p < 0.0001; NS, not significant.