Fig. 1.
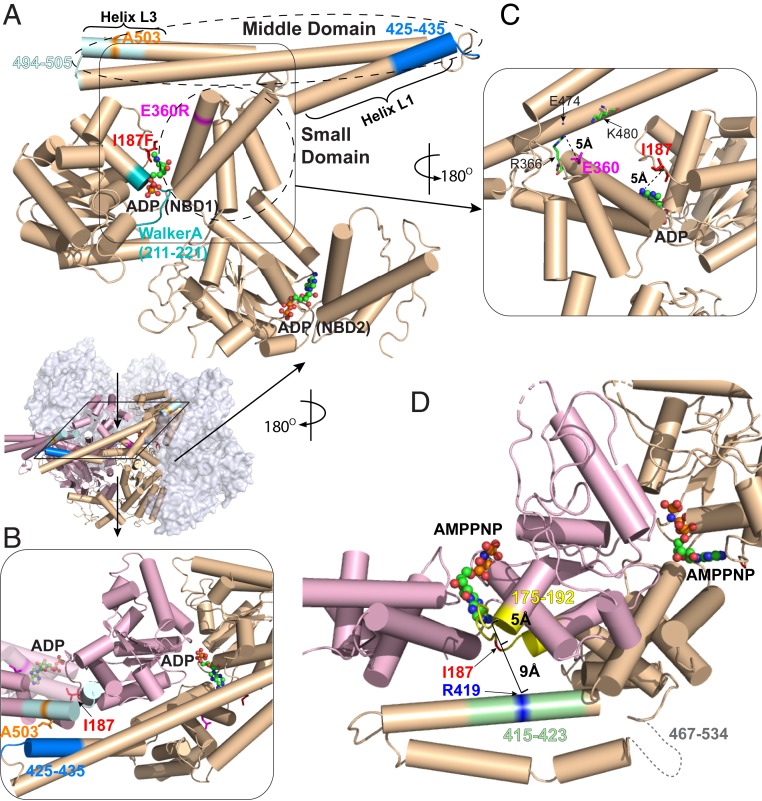
Location of potentiating Hsp104 mutations. (A) The three mutations studied here (A503S, E360R, I187F) are highlighted in stick representation along with other pertinent elements on a protomer (P3) in the ADP-bound open hexamer conformation (PDB ID code 5VY8) (6). Peptide fragments measured by HX MS are 494–505 covering helix L3 (light cyan), 425–435 covering a part of helix L1 (blue), and 211–221 covering Walker A (cyan). ADP is in ball-and-stick representation. The boundaries of the MD and the small domain of NBD1 are outlined. (B) Top view. A503 in MD helix L3 (light blue) interacts with L1 (dark blue) across an interprotomer interface (P3 in pink to P4 in wheat). Relevant structural elements are in the same colors as in A. (C) The NBD1 small domain. E360 forms a salt bridge with R366 (∼5 Å) and is close to other charged residues (e.g., K480 and E474) on the MD of the same protomer. (D) The main chain atoms of I187 are within 5 Å of the adenosine moiety of the bound nucleotide to one side and 9 Å from R419 (blue) across an interprotomer interface to the other side (P3 to P4). Segment 175–192 (interhelix B2-B3 loop) is in yellow and 415–423 in helix L1 is in light green. Bound AMPPNP is in ball-and-stick representation. The AMPPNP-bound cryo-EM structure (PDB ID code 5KNE) is used here to represent the ATP-bound state because the same MD configuration has been observed in a subclass of the ATPγS-bound protein (6, 27).