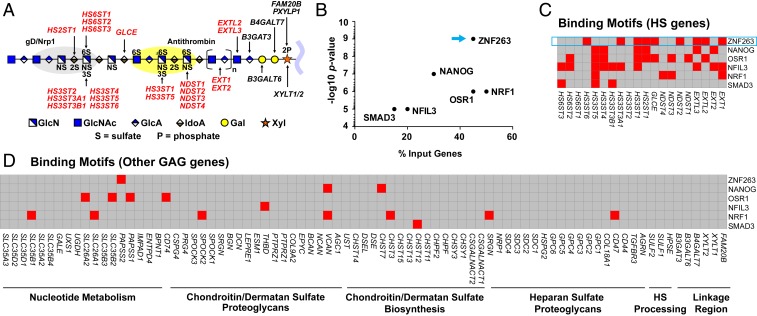
Fig. 1.
Heparin/HS structure, assembly, and regulation. (A) HS and heparin assemble while attached via a linkage tetrasaccharide to a core protein of a proteoglycan. NDSTs, HS2ST, HS3STs, and HS6STs install sulfate groups at specific sites along the HS/heparin chain, and an epimerase (GLCE) converts d-glucuronic acid to l-iduronic acid. The chain is rendered according the Symbol Nomenclature for Glycans (47). The gray and yellow oval shapes depict protein-binding sites for gD/NRP1 and antithrombin, respectively. Input genes for HOMER are indicated in bold red font. (B) HOMER motif enrichment analysis revealing TFs with predicted binding motifs in a set of HS biosynthesis genes. (C) Heatmap showing the presence (red) or absence (gray) of TF-binding motifs (y-axis) in regulatory regions of HS biosynthesis genes (x-axis), as predicted by HOMER. (D) Heatmap showing the presence (red) or absence (gray) of the same TF-binding motifs in a list of genes involved in nucleotide sugar and sulfate metabolism, CS/DS proteoglycans, CS/DS biosynthesis, HS proteoglycans, extracellular HS processing enzymes, and enzymes that generate the common linkage tetrasaccharide in heparin/HS and CS/DS.