Figure 4.
Analysis of FL Env in Lipid Bicelles and Nanodiscs in Complex with MPER-Targeting Fabs
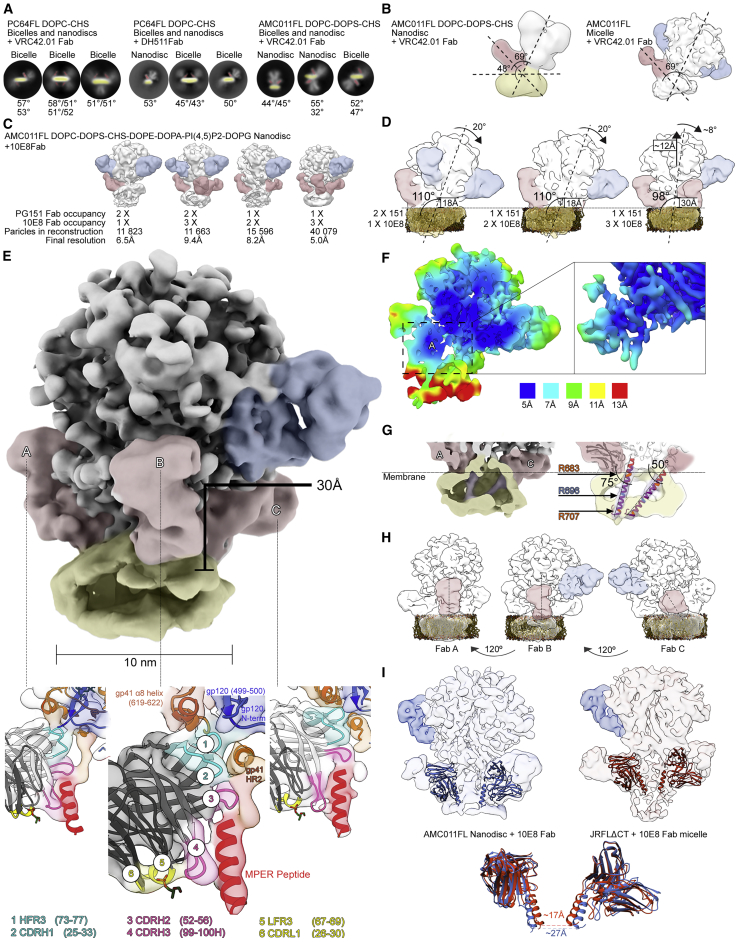
(A) Representative 2D class averages from negative-stain EM data from lipid assemblies of PC64FL and AMC011FL in complex with MPER-targeting antibodies. In all bilayer-assembled Env-MPER antibody complexes, Env was tilted at varying degrees and displaced to the edge of the bilayer. The degree of the Fab binding angle is estimated in relation to the bilayer from the given 2D class average. The lipid bilayer is highlighted in yellow, and MPER Fab is shown in red.
(B) Comparison of the VRC42.01 Fab-binding angle in the nanodisc and micelle. The angle between the Fab and the ectodomain was identical, while in the nanodisc, an additional angle can be measured between the bilayer and the Fab. PGT151 Fab is highlighted in blue throughout the figure.
(C) Low-pass filtered cryo-EM reconstructions of the AMC011FL nanodisc in complex with 10E8 Fab with different Fab occupancies. The highest resolution and particle count were obtained with the complex containing one copy of PGT151 Fab and three copies of 10E8 Fab, which is used for (E)–(I).
(D) Low-pass-filtered reconstructions with one, two, or three copies of 10E8 Fab showing degree of tilting and distance of Env from bilayer in different 10E8 Fab occupancy states.
(E) Highest resolution reconstruction and epitopes of the three Fabs with 10E8 Fab crystal structure (PDB: 5T80) docked in. Epitope components are highlighted as indicated in the panel below. The distance from the bilayer surface is also indicated.
(F) Local resolution estimation showing up to 5 Å resolution in the ectodomain and stabilized 10E8 Fab epitope.
(G) Fab A and Fab C stabilized TMD orientations are highlighted in purple. Residues marking the outer (R683) and inner (R707) bilayer surfaces are highlighted in orange and R696 in blue, which marks the crossing point of TMDs in micelle samples and is now separated. The angles of the two TMDs are estimated in relation to the bilayer surface.
(H) Two Fab orientations and dependency of Fab C on the PGT151 position are highlighted in red in low-pass-filtered maps.
(I) Comparison of 10E8 Fab docking into the AMC011FL nanodisc and JRFLΔCT micelle reconstructions (EMD-3312) showing an ∼10 Å change in the distance between the MPER peptides (residue Q135 in PDB: 5T80).
See also Figures S2, S4, and S6 and Table S1.