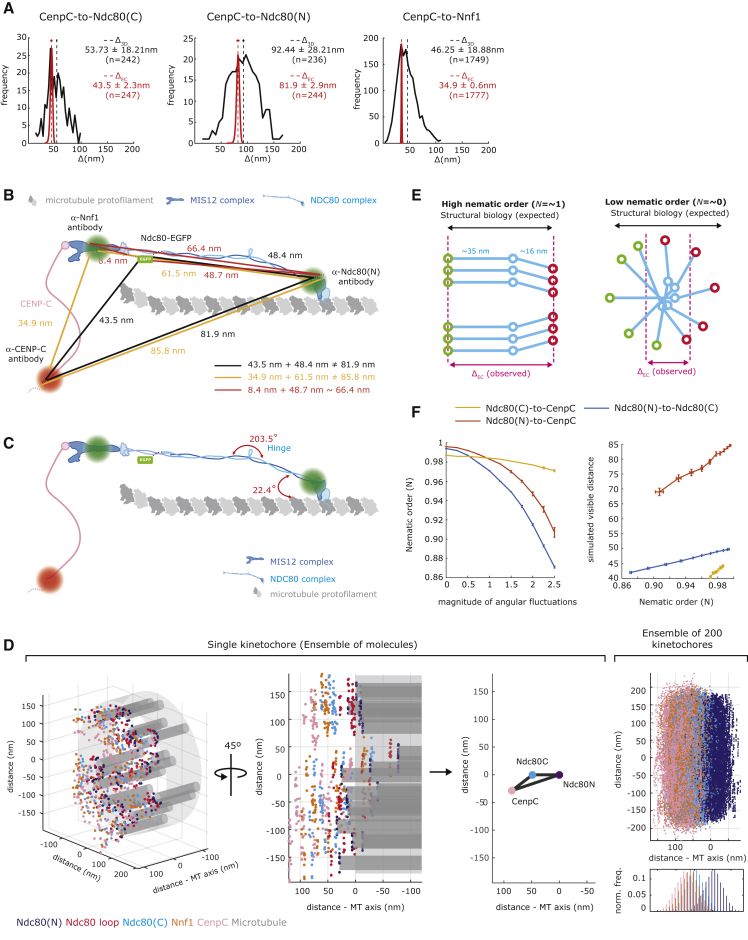
Figure 2.
Simulations of the Architecture of a Bi-oriented Kinetochore
(A) Histograms of Δ3D and ΔEC distances for CenpC to Ndc80(C) and CenpC to Ndc80(N) (Ndc80-EGFP cells) and for CenpC to Nnf1, mean (dashed line), and SD (horizontal bar) values are indicated at right.
(B) Schematic showing the inferred architecture of CenpC-MIS12-NDC80. Approximate antibody binding sites (or EGFP tagging) are indicated with green dots. Lines indicate ΔEC mean values (dotted lines show SDs) obtained in 3-fluorophore experiments: CenpC-Ndc80(C)-Ndc80(N) (black), CenpC-Nnf1-Ndc80(N) (orange), and Nnf1-Ndc80(C)-Ndc80(N) (red).
(C) Schematic of CenpC-MIS12-NDC80 showing best-fitted angles (red) from simulations; see Methods S1.
(D) Simulated kinetochore organization. Left and center left: 2 perspectives of the same simulated kinetochore; each dot represents a single protein at the specified position (labeled by color). Center right: averages of the simulated position of each marker shown in the plane of the CenpC-Ndc80(N)-Ndc80(C) triangle. Right: an ensemble of 200 simulated kinetochores aligned along their microtubule axis and to the mean of their Ndc80(N) markers. Histogram of the marker density along K-fiber for the above plot.
(E) Schematic representation of NDC80 complexes under high (N = ~1) and low (N = ~0) nematic order. Circles represent Ndc80(C) (green), Ndc80 loop (blue), and Ndc80(N) (red).
(F) Left: change in the nematic order of the NDC80 complex population with respect to the magnitude of Ndc80 loop stochastic angular fluctuations. Right: average distance between the indicated molecular markers against the nematic order. Simulations based on 200 kinetochores.