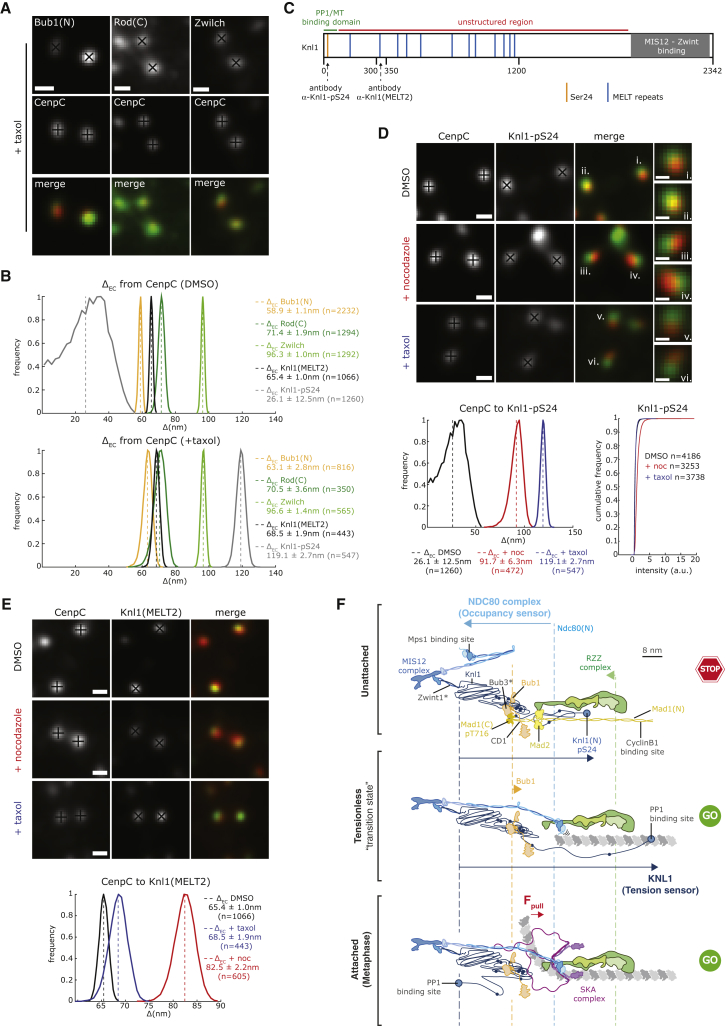
Figure 7.
Knl1 1–300 Unravels upon the Loss of Kinetochore Tension
(A) Kinetochore pairs stained with anti-Bub1, anti-Rod, and anti-Zwilch antibodies in cells treated with 1 μM taxol for 15 min. Scale bars, 500 nm.
(B) Histograms of the ΔEC distances from CenpC to Bub1, Rod, Zwilch, Knl1-MELT2, and pKnl1(S24) in DMSO and taxol. Mean (dashed line) and SD values are indicated at right.
(C) Schematic map of Knl1 where the positions of Serine24 (Ser24, orange), MELT repeats (blue), and MIS12-Zwint-binding domain (gray) are shown. Lines indicate the PP1-microtubule binding site (green) and the unstructured region (red). Arrows indicate the binding sites of anti-Knl1-pS24 and anti-Knl1(MELT2) antibodies used in this study.
(D) Top: kinetochores stained with anti-CenpC and anti-Knl1-pS24 antibodies in cells treated with 3.3 μM nocodazole for 2 h, 1 μM taxol for 15 min, or DMSO. Scale bar, 500 nm. Insets show enlargements for the indicated kinetochores. Scale bar, 250 nm. Bottom: histograms of the ΔEC between CenpC and Knl1-pS24 in cells treated as above. Mean (dashed line) and SD values are indicated below. Cumulative frequency plots display the Knl1-pS24 intensity in DMSO-, nocodazole (+noc)-, and taxol-treated cells. The Knl1-pS24 signal is normalized to the CenpC signal. Signals are background subtracted.
(E) Top: kinetochores stained with anti-CenpC and anti-Knl1(MELT2) antibodies and treated as in (D). Scale bars, 500 nm. Bottom: histograms of the ΔEC distances between CenpC and Knl1 in DMSO-, nocodazole (+noc)-, and taxol-treated cells. Mean (dashed line) and SD values are indicated at right.
(F) Schematics representing the ensemble average arrangement of MIS12 complex (dark blue), NDC80 complex (light blue), Bub1 (orange), RZZ complex (green), Mad1:Mad2 (yellow), Knl1 (black), and SKA (purple) in RPE1 cells treated with nocodazole (top), taxol (center), and DMSO (bottom). A single microtubule protofilament is shown in gray and dotted lines represent the position of the indicated complexes in control cells. The arrows indicate the change in position between different conditions. The positions of proteins marked with an asterisk are inferred from known biochemical data. These kinetochore organizational states can be integrated with known checkpoint and error-correction mechanisms as follows: at unattached kinetochores (top), NDC80 is disordered and in an auto-inhibited state, while the first 300 aa of Knl1 are in an extended conformation. Aurora B kinase activity dominates, kinetochore substrates (i.e., Ndc80 and Knl1) are phosphorylated, and Ndc80 is in a low-affinity microtubule-binding state. We note that Mad1 can bridge from the outer kinetochore to the corona, with interactions at either end necessary for stable binding and spindle checkpoint activation. When end-on attachment forms (center), but no tension is generated (e.g., monotelic attachments in which one sister is attached and the other is unattached), the checkpoint is silenced on the attached sister concomitantly with the straightening of Ndc80, which binds to the microtubule lattice. As force from the microtubule depolymerization generates tension (bottom), SKA complexes are recruited and the Knl1 amino terminus ravels. PP1 now fully binds, leading to kinetochore dephosphorylation (counteracting Aurora B) and increased microtubule binding affinity. This model can also explain error correction following the loss of tension at a bi-oriented kinetochore, which would switch the system into the transition (center panel) state (see Discussion for details).