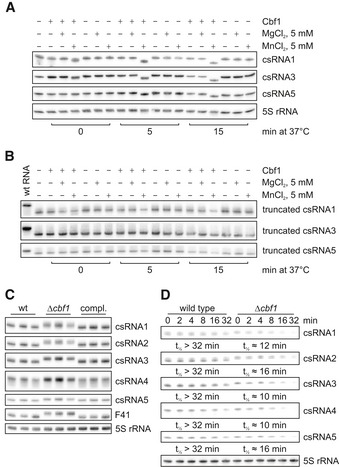
Figure 6. Cbf1 acts as an sRNA‐stabilizing 3′→5′ exonuclease.
-
AIn vitro RNase assay. In vitro‐transcribed RNAs were incubated in the presence or absence of Cbf1, MgCl2, and MnCl2. Reactions were stopped at the indicated time points and the RNA run on denaturing 10% PAGE followed by northern blotting. All csRNAs, but not 5S rRNA, are cleaved in the presence of Cbf1 and MnCl2.
-
BIn vitro RNase assay with truncated RNAs. The setup of the experiment was the same as in A, the difference being that the in vitro‐transcribed RNAs were missing the single‐stranded uridine overhang 3′ of the Rho‐independent terminator. Wild‐type (wt) RNA used in A was loaded as reference. No cleavage can be observed in any of the conditions.
-
CNorthern blots of denaturing 10% PAGE of total RNA extracted from wild type (wt), Δcbf1 and the complementation (compl.) strain. Upon deletion of cbf1, an upshift of the tested ncRNAs, but not of 5S rRNA is visible.
-
DIn vivo RNA stability assays. After blocking transcription with rifampicin, RNA was collected at the indicated time points and visualized using northern blotting of 6% denaturing PAGE. Interestingly, knockout of cbf1 increases stability of the tested RNAs. 5S rRNA was used as loading control. RNA half‐lives were calculated based on at least two independent experiments (Fig EV5B–F).
Source data are available online for this figure.
