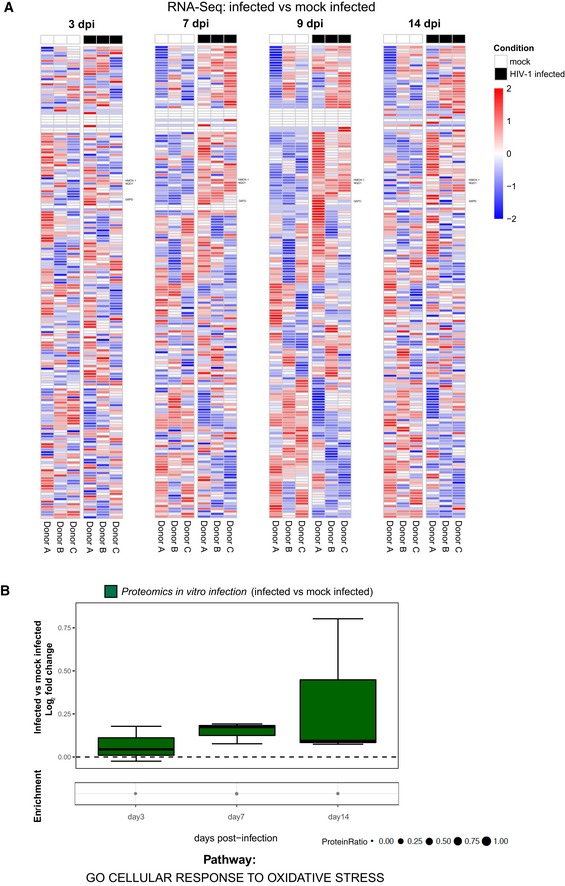
Figure EV2. Influence of HIV‐1 replication on antioxidant gene and protein expression.
-
A, BRNA‐Seq (A) and proteomic (B) analyses of the relative expression over time of antioxidant genes and proteins in primary CD4+ T cells infected in vitro with HIV‐1 or mock infected. (A) Heatmaps of the standardized expression of antioxidant genes in HIV‐1‐infected and mock‐infected samples over time. Expression levels were standardized [(mean gene expression − SD)/SD] for each gene in each time point. Genes considered for further analysis in the paper are named on the right. (B) Boxplots of proteomics data illustrating the log2 fold change expression for each time point in infected as compared to mock‐infected cells. Median and 25–75 percentiles are depicted, while whiskers extend from the hinge to the highest or lowest value that is within 1.5 * IQR (inter‐quartile range). Data beyond the end of the whiskers are outliers and plotted as points. Data were analyzed by Fisher test, for each time point, dots illustrate the pathway enrichment analysis of proteins up‐regulated in infected vs. matched mock‐infected controls. Dots size indicates the fraction of differentially expressed proteins in the pathway. All analyses were conducted using the pathway GO cellular response to oxidative stress (184 genes; GO:0034599). Number of donors = 3 biological replicates.
