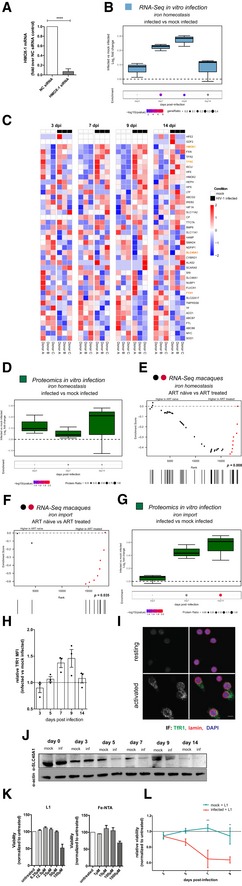
Figure EV4. Expression of iron homeostasis and import markers and effect of iron chelation on cell viability in HIV‐1 infection.
-
AHMOX‐1 mRNA expression in Jurkat‐TAg cells transfected with non‐targeting siRNA or siRNAs targeting the HMOX‐1 gene. Cells were collected 72 h post‐transfection and analyzed by qPCR. Data (mean ± SEM; n = 3 technical replicates) were normalized using the NC control and analyzed by unpaired t‐test. ****P < 0.0001.
-
B–GRNA‐Seq (B, C, E, F) and proteomic (D, G) analysis of different time points of primary CD4+ T cells infected in vitro with HIV‐1 or mock infected (B, C, D, G) and PBMCs of macaques infected with SIVmac239 before and after suppression of viremia with ART (E, F). The gene sets used for the analyses are GO cellular ion iron homeostasis (46 genes, GO:0006879; B–E) and GO iron ion import (12 genes, GO:0097286; F, G). Data were analyzed by Fisher test (B, D, G; number of donors = 3 biological replicates) or GSEA (E, F, number of animals = 8). Boxplots in (B, D, G) illustrate the log2 fold change expression for each time point in infected as compared to mock‐infected cells. Depicted in each boxplot are the median and 25–75 percentiles, while whiskers extend from the hinge to the highest or lowest value that is within 1.5 * IQR (inter‐quartile range) of the hinge. Data beyond the end of the whiskers are outliers and plotted as points. Dots illustrate the pathway enrichment analysis of proteins up‐regulated in infected cells. Dots are color‐coded based on the enrichment q‐values, and their size indicates the fraction of differentially expressed proteins in the pathways; gray dots are not statistically significant. Heatmaps in (C) depict the standardized expression of genes of the GO cellular ion iron homeostasis pathway in HIV‐1‐infected and mock‐infected samples over time. Expression levels were standardized [(mean gene expression − SD)/SD] for each gene in each time point. Genes considered for further analysis in the paper are highlighted. Symbols in the GSEA enrichment plots (E, F) represent the position of the gene set members in the transcriptome ranked by differential expression between ART‐naïve and ART‐treated time points. Red color indicates leading edge genes.
-
HRelative TfR1 expression over time in HIV‐1‐infected vs. mock‐infected CD4+ T cells as measured by flow cytometry. Data are expressed as relative MFI (median fluorescence intensity) of TfR1 in HIV‐1‐infected vs. mock‐infected cells (mean ± SEM; n = 3 biological replicates).
-
IRepresentative immunofluorescence image of TfR1 expression in CD4+ T cells resting or activated for 72 h with α‐CD3/CD28 beads. Scale bar = 10 μm.
-
JProtein expression over time of SLC40A1 in CD4+ T cells infected with HIV‐1 or mock infected as assessed by Western blot.
-
KRelative viability of CD4+ T cells incubated for 48 h with various concentrations of the iron chelator deferiprone (L1) or the iron donor ferric nitriloacetate (Fe‐NTA). Viability was measured using the MTT assay. Absorbance values were normalized over untreated controls and expressed as percentage (mean ± SEM, n = 3 biological replicates).
-
LTime course of the relative viability of HIV‐1‐infected or mock‐infected CD4+ T cells left untreated or treated with 50 μM L1. Viable cells were identified by flow cytometry through FSC/SSC gating (mean ± SEM, number of donors n = 3). **P < 0.01; ***P < 0.001.
Source data are available online for this figure.
