Figure 1.
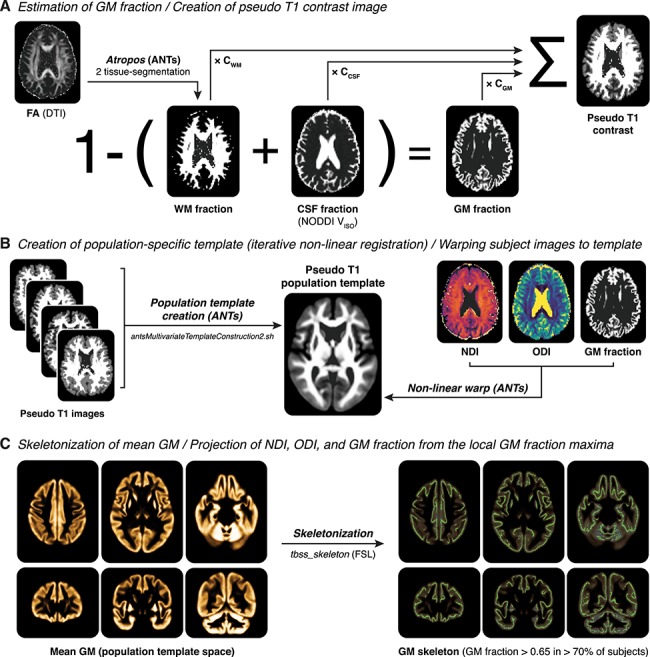
Processing steps of GBSS. (A) For each subject, a gray matter (GM) fraction map is generated by subtracting the white matter (WM) fraction (estimated from the subject’s FA map using Atropos) and the CSF fraction (estimated from the subject’s NODDI VISO parameter map) from 1. The GM, WM, and CSF fraction are then used to generate a pseudo T1-weighted contrast image. (B) Pseudo T1-weighted images from all subjects are used to create a population template via iterative nonlinear registration in ANTs (antsMultivariateTemplateConstruction2.sh). The warp fields generated from this step are then used to nonlinearly warp NDI, ODI, and GM fraction images for each subject into population template space. (C) GM fraction images in population template space are averaged to generate a mean GM image, which is then skeletonized using FSL’s tbss_skeleton tool. Finally, NDI and ODI are projected onto the GM skeleton from the local GM fraction maxima, and the GM skeleton is thresholded to only include voxels with GM fraction > 0.65 in > 70% of participants.
