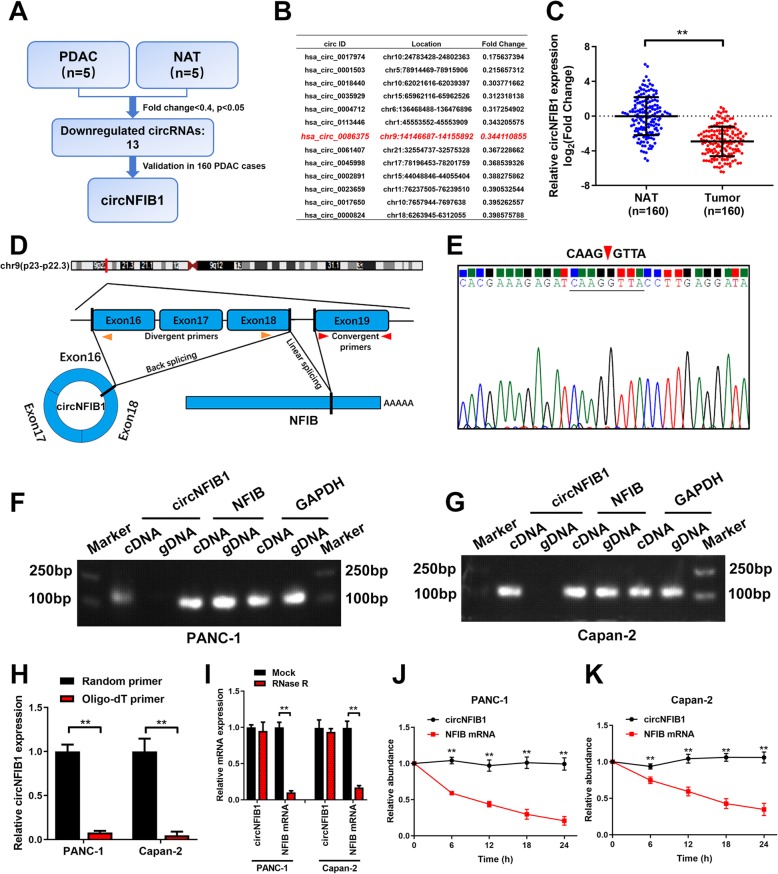
Fig. 1.
The identification and characterization of circNFIB1 in PDAC. a Schematic illustration of the identification of circRNAs downregulated in PDAC tissues compared with NATs. b The downregulated circRNAs in PDAC identified from NGS were shown. c qRT-PCR analysis of circNFIB1 expression in PDAC tissues (n = 160) paired with NATs (n = 160). The nonparametric Mann-Whitney U test was used to compare different groups. d Schematic illustrations showed the genomic loci of circNFIB1. circNFIB1 was produced by exons 16 to 18 of NFIB. e The back-splice junction of circNFIB1 was identified by Sanger sequencing. f and g PCR analysis for circNFIB1 and NFIB in the cDNA and gDNA of PANC-1 (f) or Capan-2 (g) cells. GAPDH was used as NC. h qRT-PCR analysis of circNFIB1 expression using random primers or oligo-dT primers. i circNFIB1 and NFIB expression in PDAC cells treated with or without RNase R was assessed by qRT-PCR. j and k qRT-PCR analysis of circNFIB1 and NFIB mRNA in PANC-1 (j) or Capan-2 (k) cells treated with actinomycin D at the indicated time points. Significance level was assessed using two-tailed Student t-tests. Figures with error bars showed the standard deviations of three independent experiments. *p < 0.05 and **p < 0.01