Figure 2. Intact OXPHOS Promotes ER-phagy.
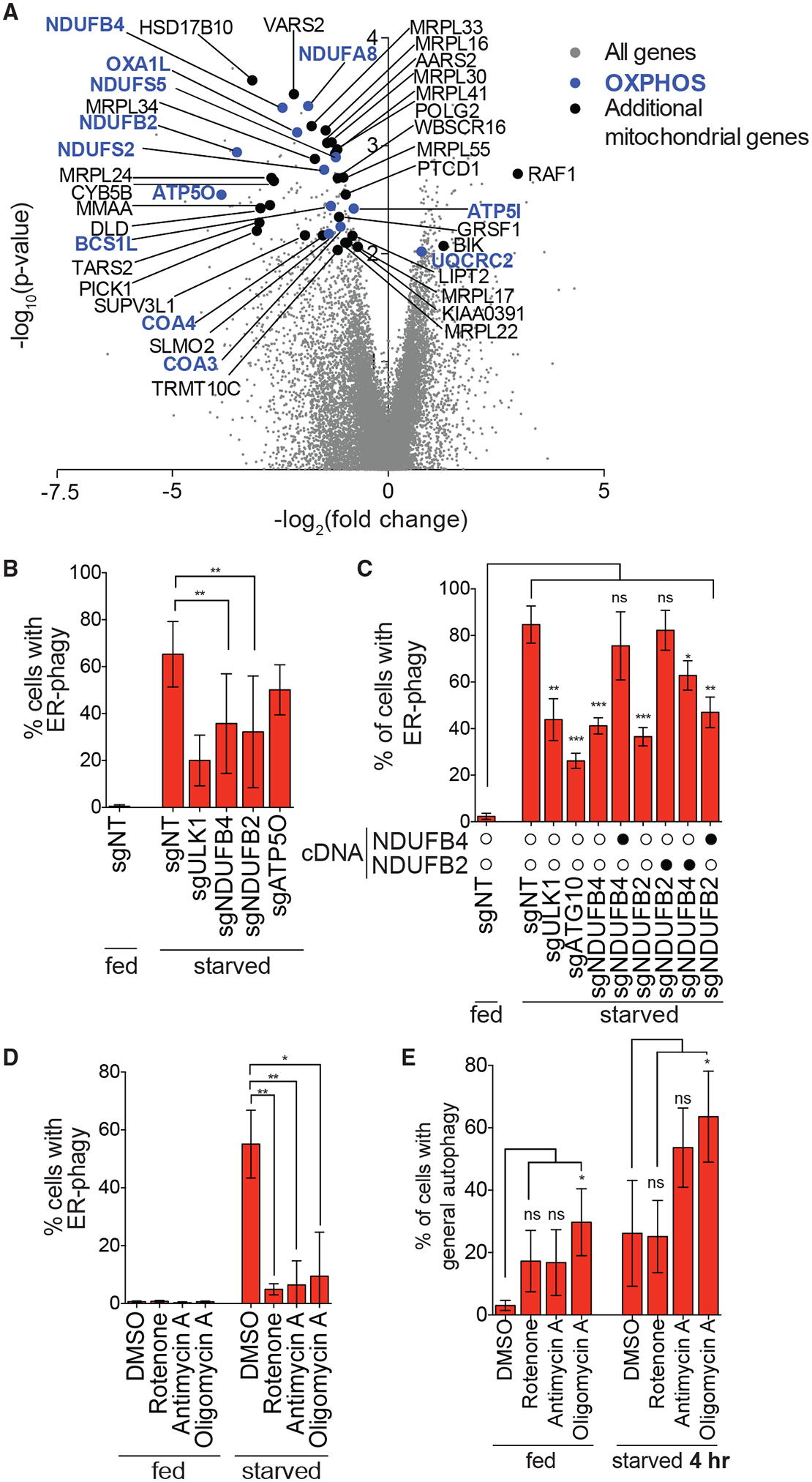
(A) Genes that are components of the OXPHOS pathway were top hits in screen (highlighted in blue). Additional mitochondria-related genes are indicated in black and all other targeting sgRNAs are indicated in gray.
(B) Knockdown of NDUFB4 and NDUFB2 significantly inhibit ER-phagy. HCT116 CRISPRi EATR cells were transduced with sgRNAs targeting ULK1, NDUFB4, NDUFB2, or ATP5O and starved for 16 h before FACS measurement for ER-phagy. Data are presented as mean ± SD of eight biological replicates.
(C) Re-expression of cognate cDNA of each OXPHOS gene rescues ER-phagy. HCT116 CRISPRi EATR cells were transduced with NDUFB4 or NDUFB2 cDNA constructs and then transduced with the indicated sgRNAs. Cells were starved for 16 h before FACS measurement for ER-phagy. Data are presented as mean ± SD of three biological replicates.
(D) Small-molecule inhibitors of the different OXPHOS compartments phenocopy the effect of genetic inhibitions. HCT116 EATR cells were treated with rotenone, antimycin A, or oligomycin A and starved for 16 h before FACS measurement of ER-phagy. Data are presented as mean ± SD of three biological replicates.
(E) General autophagy proceeds in cells where NDUFB2, NDUFB4, or ATP5O are knocked down. HCT116 CRISPRi cells expressing mCherryeGFP-LC3B were transduced with the indicated sgRNAs. Cells were starved for 4 h before FACS measurement for general autophagy. Data represent mean ± SD of four biological replicates.
See also Figures S2 and S3.
