Figure 5. DDRGK1 Recruits UFL1 to the ER Surface via the PCI Domain.
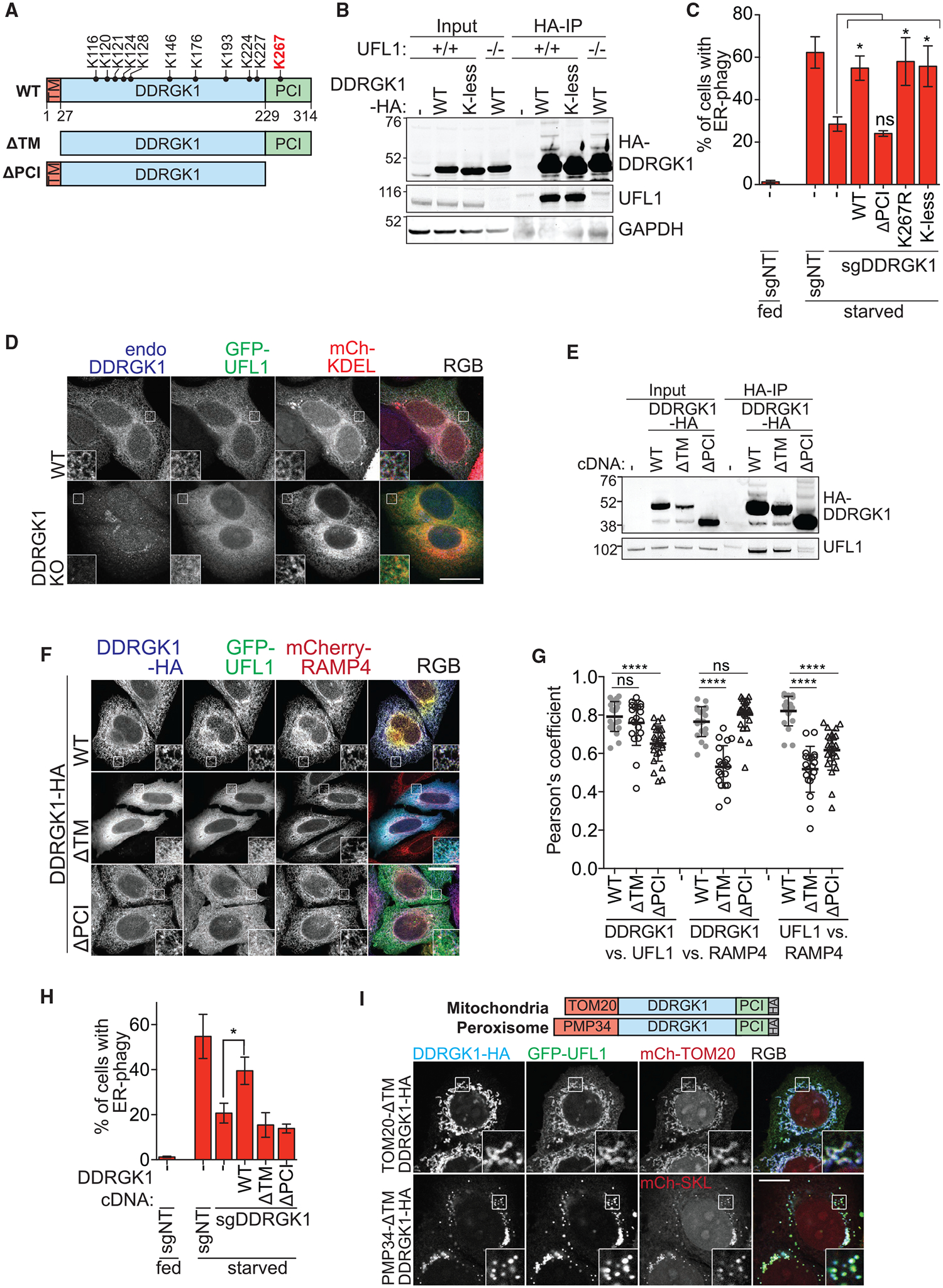
(A) Schematic of DDRGK1 domains and its conserved lysine residues. The reported major lysine residue for UFMylation (K267) is labeled in red. The two truncated forms of DDRGK1 that either lacks the N-terminal transmembrane domain (ΔTM) or the C-terminal proteasome component domain (ΔPCI) are also shown.
(B) Post-translational modification of DDRGK1 occurs on lysine residues. Parental or UFL1 knockout HCT116 cells were transfected with either wild-type (WT) or lysine-less (K-less) DDRGK1-HA constructs. Cells were harvested for HA immunoprecipitation.
(C) DDRGK1’s role during ER-phagy does not require post-translational modification on any lysine residue. HCT116 CRISPRi EATR cells were transduced with DDRGK1 sgRNA and then rescued using the indicated DDRGK1-HA mutant constructs. Cells were starved for 16 h before FACS ER-phagy measurement. Data represent mean ± SD of three biological replicates.
(D) Loss of DDRGK1 relocalizes UFL1 to the cytoplasm. Wild-type or DDRGK1KO HeLa cells were transiently transfected with GFP-UFL1 and mCherry-KDEL for 48 h. Cells were then fixed and immunostained for endogenous DDRGK1. Insets represent a 3-fold enlargement of boxed areas. Scale bar represents 20 μm.
(E) DDRGK1 interacts with UFL1 via its PCI domain. Parental HCT116 cells were stably transfected with the indicated DDRGK1-HA mutant constructs. Cells were then harvested for HA immunoprecipitation.
(F) DDRGK1 recruits UFL1 to the ER. DDRGK1KO HeLa cells were stably transduced with mCherry-RAMP4 (ER marker) and the indicated DDRGK1-HA mutant constructs. Cells were then transiently transfected with GFP-UFL1 for 24 h. Cells were then fixed and immunostained for HA epitope. Representative images are shown. Insets represent a 3-fold enlargement of boxed areas. Scale bar represents 10 μm.
(G) Pearson’s correlation coefficient for (F) was measured between DDRGK1 versus UFL1, DDRGK1 versus RAMP4, and UFL1 versus RAMP4. Data were generated from one biological experiment, and 20–26 cells were analyzed from each condition.
(H) DDRGK1’s role during ER-phagy requires both the SP and PCI domains. HCT116 CRISPRi EATR cells with DDRGK1 knockdown were rescued using the indicated DDRGK1-HA mutant constructs. Cells were then starved for 16 h and ER-phagy was measured by FACS analysis. Data represent mean ± SD of three biological replicates.
(I) DDRGK1’s determines the subcellular localization of UFL1. DDRGK1 knockout HeLa cells were stably transduced with either TOM20MTS-DDRGK1-dSP-HA (MTS, mitochondrial targeting signal) or PMP34-DDRGK1-dSP-HA and transiently transfected with GFP-UFL1 and the respective mCherry-organelle constructs. Insets represent a 3-fold enlargement of boxed areas. Scale bar represents 10 μm. See also Figure S5.
