Figure 7. UFMylation-Mediated ER-phagy Represses IRE1α UPR.
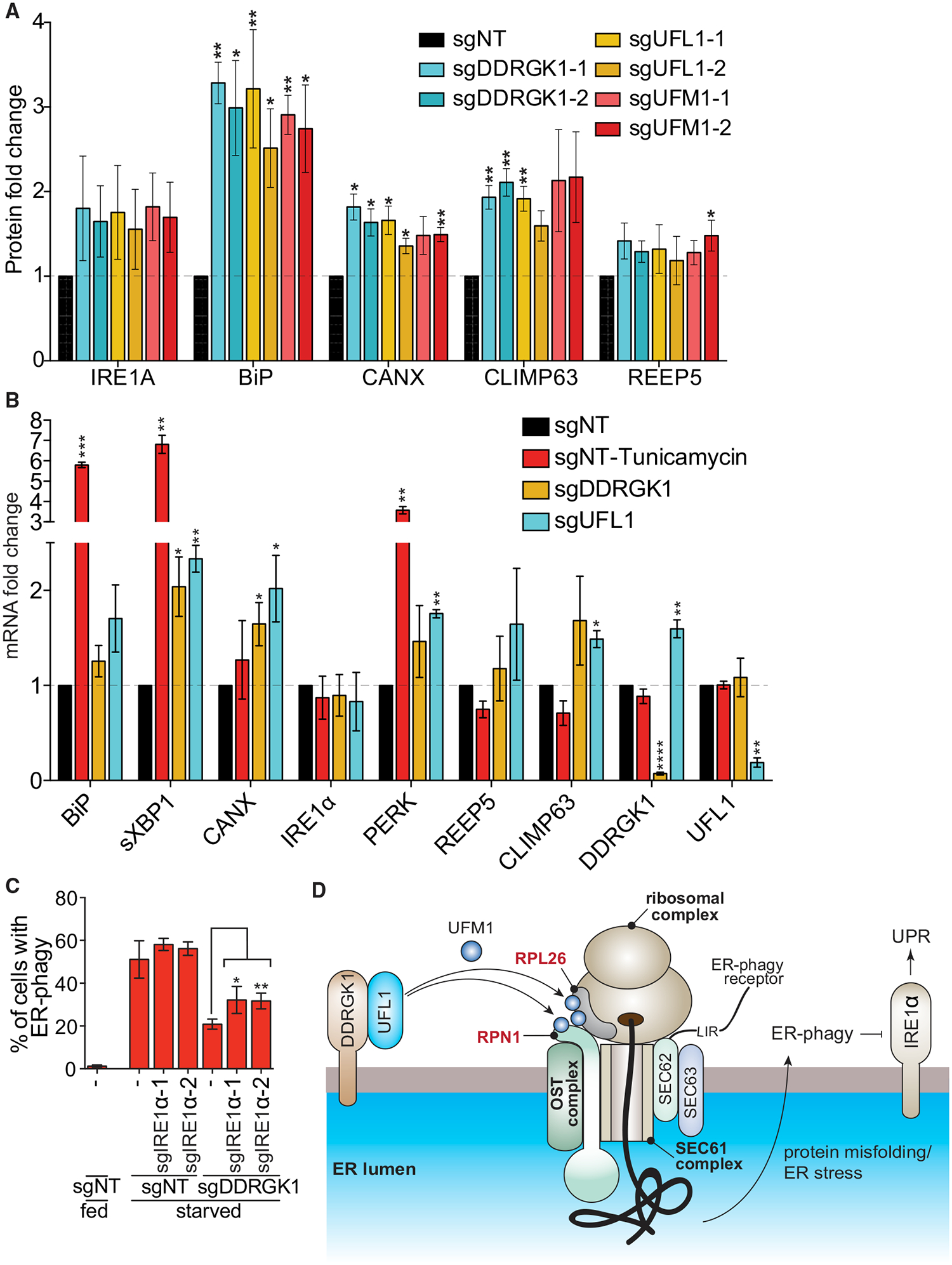
(A) Dysregulation of UFMylation results in upregulation of UPR. HCT116 cells were transduced with the indicated sgRNAs and harvested for western blotting analysis. The graph represents densitometry measurement of the indicated proteins upon sgRNA knockdown. A representative blot is shown in Figure S7B. Data represent mean ± SD of three biological replicates.
(B) Dysregulation of UFMylation transcriptionally upregulates UPR markers except IRE1 α. HCT116 CRISPRi cells were transduced with the indicated sgRNAs. Tunicamyin (0.5 μg/mL; 4 h) was used as a positive control for ER stress. Cells were harvested for qRT-PCR measurement of the indicated ER or UPR genes. Data represent mean ± SD of three biological replicates.
(C) Knockdown of IRE1a partially restores ER-phagy in DDRGK1-depleted cells. HCT116 CRISPRi EATR cells transduced with the indicated sgRNAs and starved for 16 h before FACS measurement for ER-phagy. Data represent mean ± SD of three biological replicates.
(D) Proposed model for the role of UFMylation during ER-phagy. DDRGK1 acts as an ER surface adaptor that recruits UFL1. At least two ER surface proteins that are in close proximity, RPN1 and RPL26, are UFMylated during ER-phagy. Dysregulation of UFMylation inhibits ER-phagy, and this potentially results in accumulation of ER stress and subsequently activates the IRE1 a-mediated unfolded protein response pathway.
See also Figure S7.
