Extended Data Fig. 10. Circadian rhythms in excised roots at various temperatures.
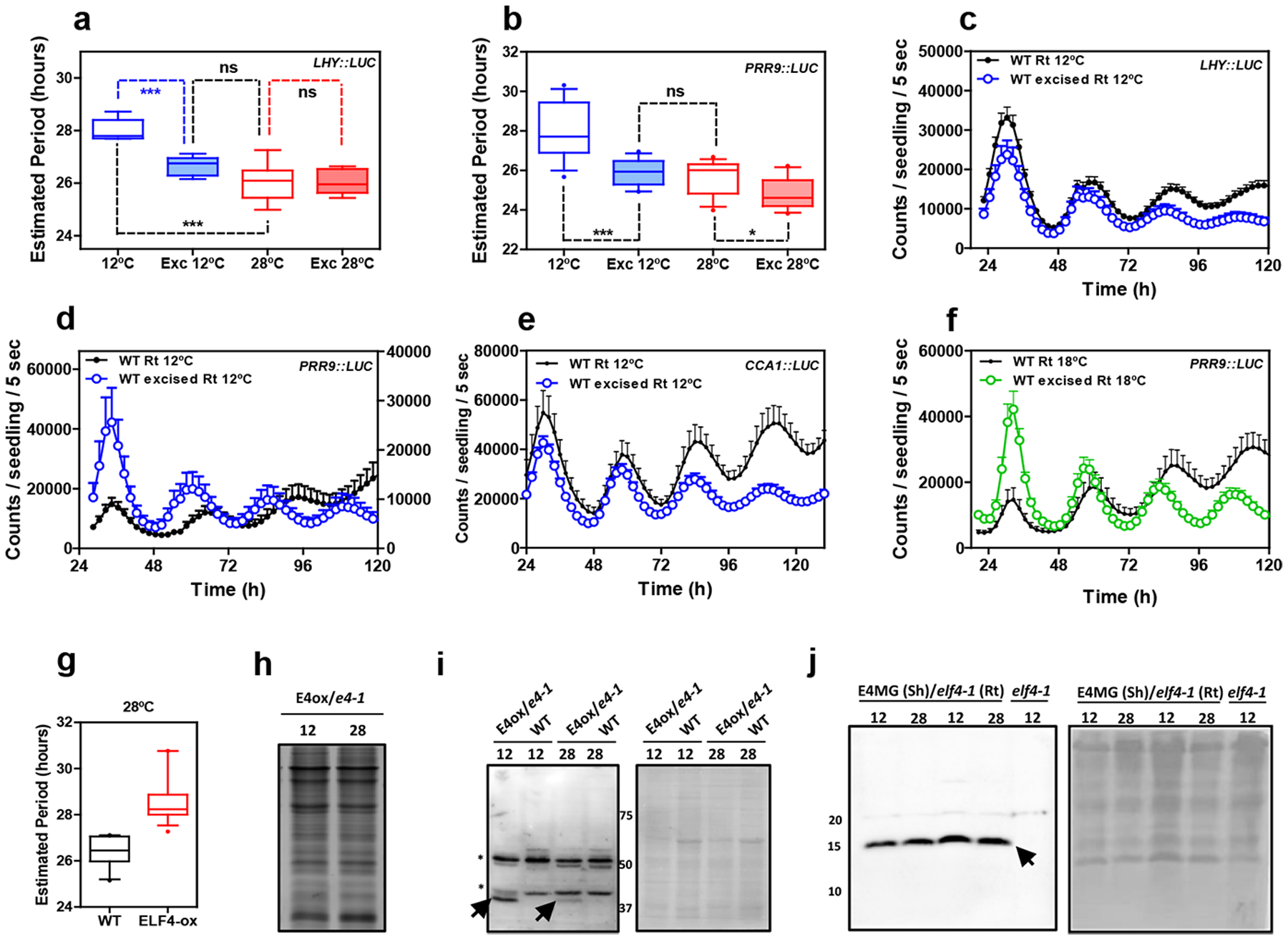
Circadian period estimates of a, LHY::LUC in intact (n=8) versus excised WT roots (n=7) at 12°C and 28°C (n=8 for intact and n=8 for excised) and b, PRR9::LUC in intact (n=12) versus excised WT roots (n=13) at 12°C and 28°C (n=14 for intact and n=16 for excised). Data are represented as the median ± max and min; 25–75 percentile. *** p-value<0.0001; * p-value<0.05; ns: non-significant p=0.369; two-tailed t-tests with 95% of confidence. Luminescence rhythmic oscillation in WT intact and excised roots (n=8 for each) at 12°C of c, LHY::LUC, d, PRR9::LUC (n=8 for each) and e, CCA1::LUC (n=4 for excised, n=5 for intact). f, Luminescence of PRR9::LUC rhythmic oscillation in WT intact and excised roots at 18°C (n=16 for each). c-f, Data are represented as the means + SEM. g, Circadian period estimates of LHY::LUC in WT (n=12) and ELF4-ox (n=17) at 28°C. Data are represented as the median ± max and min; 25–75 percentile. Promoter activity analyses were performed under constant light conditions previous synchronization of plants under LD cycles at 22°C. h, Coomassie Blue staining of protein extracts from roots of ELF4-ox-GFP scion (E4-ox) grafted into elf4–1 rootstock (e4–1) at 12°C and 28°C. i, Western-blot analyses of ELF4-GFP protein accumulation (arrows) in roots of ELF4-ox-GFP scion (E4-ox) grafted into elf4–1 rootstock (e4–1) at 12°C and 28°C. WT protein extracts were used as a control. Asterisks denote non-specific bands. Ponceau S staining of the membrane is shown in the right panel. j, Western-blot analyses of ELF4 protein accumulation (arrow) in shoots of ELF4 Minigene (E4MG) grafted into elf4–1 rootstock (e4–1) at 12°C and 28°C. elf4–1 protein extracts were used as a control. Ponceau S staining of the membrane is shown in the right panel. The “n” values refer to independent samples. a-j, Two biological replicates were performed for all experiments, with measurements or analyses taken from distinct samples grown and processed at different times.
