Figure 1.
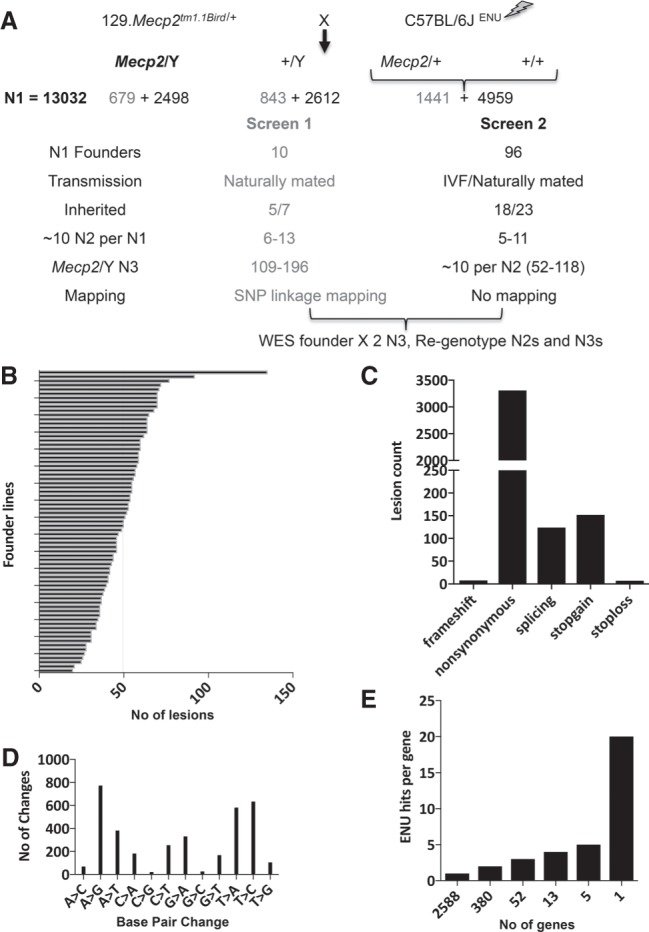
Overview of two dominant suppressor ENU mutagenesis screens in a Mecp2tm1.1Bird mouse model. (A) Male C57BL/6J male mice were treated with ENU and mated to 129.Mecp2tm1.1Bird/+ females. Numbers obtained from screen 1, already published, are in gray, and screen 2 in black. A subset of N1 male founder mice (N1) carrying the mutant Mecp2 allele and showing suppression of disease phenotypes were bred (naturally or by IVF) for inheritance. N2 offspring were mated to 129.Mecp2tm1.1Bird/+ females or 129S6/SvEvTac males to generate N3s, of which DNAs from at least two animals from different N2 families that showed trait improvement were whole-exome sequenced (WES). N3 offspring were genotyped to analyze for linkage or association. (B) The WES was analyzed for 71 founder lines, producing 3601 potentially causative variants. Variants ranged in number from 20 to 135, with a mean of 50. (C) The types of ENU-induced lesions generated were mostly nonsynonymous missense mutations (3310, 92%), followed by nonsense mutations (152, 4%) and splicing mutations (124, 3%). Frameshift mutations or the loss of a STOP codon accounted for the remainder (1%). (D) The most common types of mutations induced by ENU were A to G and T to C transition mutations (22% and 18%, respectively), closely followed by transversions (A to T [11%] and T to A [16%]), whereas the fewest were C to G and G to C transversions (<1%). (E) The majority of genes with new ENU-induced lesions were unique, with 2588 genes carrying only one mutation. A single gene, titin (Ttn), had 20 alleles.
