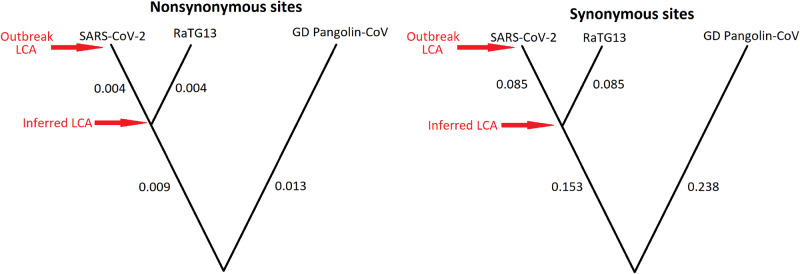
Figure 3.
Schematic phylogenetic trees, not drawn to scale, inferred from nonsynonymous (left) and synonymous sites (right) using the estimated divergence values per site from Table 1 of Tang et al. (2020), assuming clock-like mutation rates. The last common ancestor (LCA) of the SARS-CoV-2 outbreak is much closer to that of the LCA shared with the bat-infecting RaTG13 sample in nonsynonymous sites than in synonymous sites. Accession numbers from GISAID for the RaTG13 and Guandong (GD) Pangolin-CoV samples are EPI_ISL_402131 and EPI_ISL_410721, respectively.