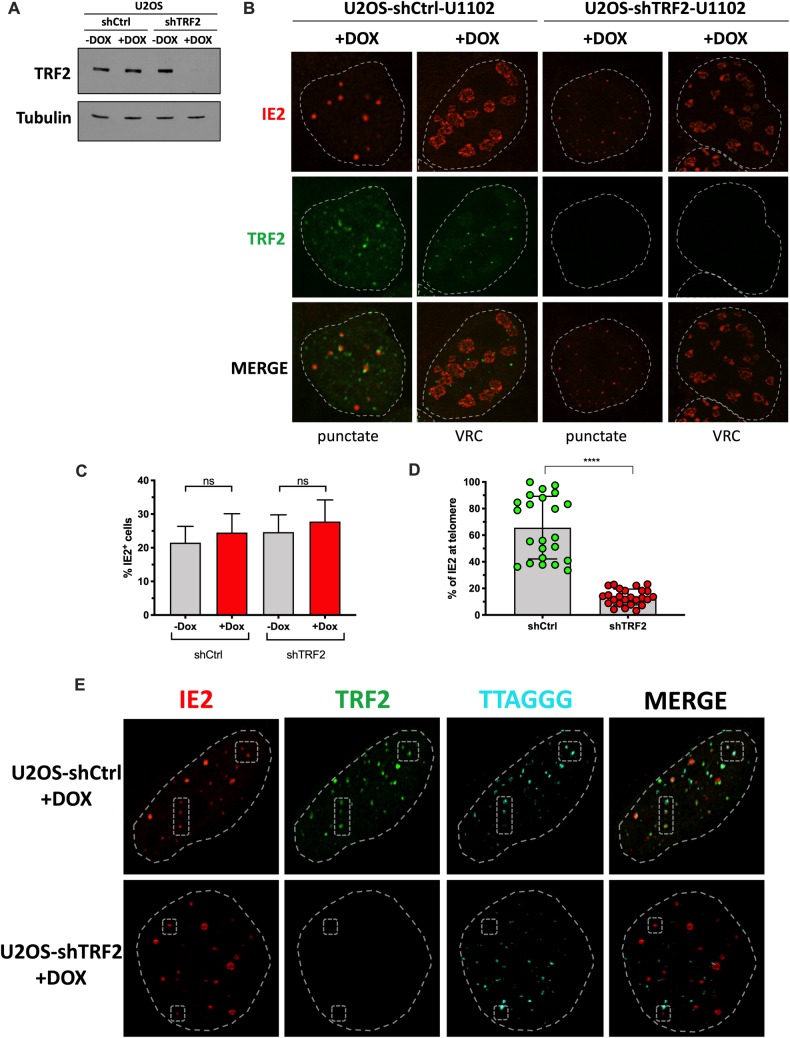
Fig 7. TRF2 is required for IE2 localization with telomeres.
U2OS cells were transduced with a lentiviral vector coding for a Dox inducible control shRNA (shCtrl) or a shRNA against TRF2 (shTRF2) and selected with puromycin +/- Dox for a week. A) Western blot analysis of TRF2 expression one week post selection. Membranes were also probed with anti-tubulin antibodies to show the input material loaded. B) One week post selection, +Dox cells were infected with HHV-6A for 48h and processed for IFA using anti-TRF2 (green) and anti-IE2 (red). Cells with IE2 in punctate form and cells with large patchy IE2, likely to represent VRC, are shown. Nuclei are outlined by dashed lines. C) The percentage of HHV-6A infected cells (from B) was estimated after counting a minimum of 700 cells and scoring the IE2+ ones. Results are expressed as mean %IE2+ cells ± SD. D) Mean percentage ± SD of IE2 localizing with telomeres in the presence (shCtrl +Dox) and absence (shTRF2 +Dox) of TRF2. Each dot represents the % of IE2 foci localizing with telomeres in one nucleus. ****p<0.0001. E) IF-FISH confocal images of shCtrl (+Dox) and shTRF2 (+Dox) cells analyzed for TRF2 (green), IE2 (red) and telomeres (cyan). Nuclei are outlined by dashed circles. Examples of IE2 localizing with telomeres (top row) or not found with telomeres (bottom row) are highlighted by the dashed polygons.