Fig. 12.
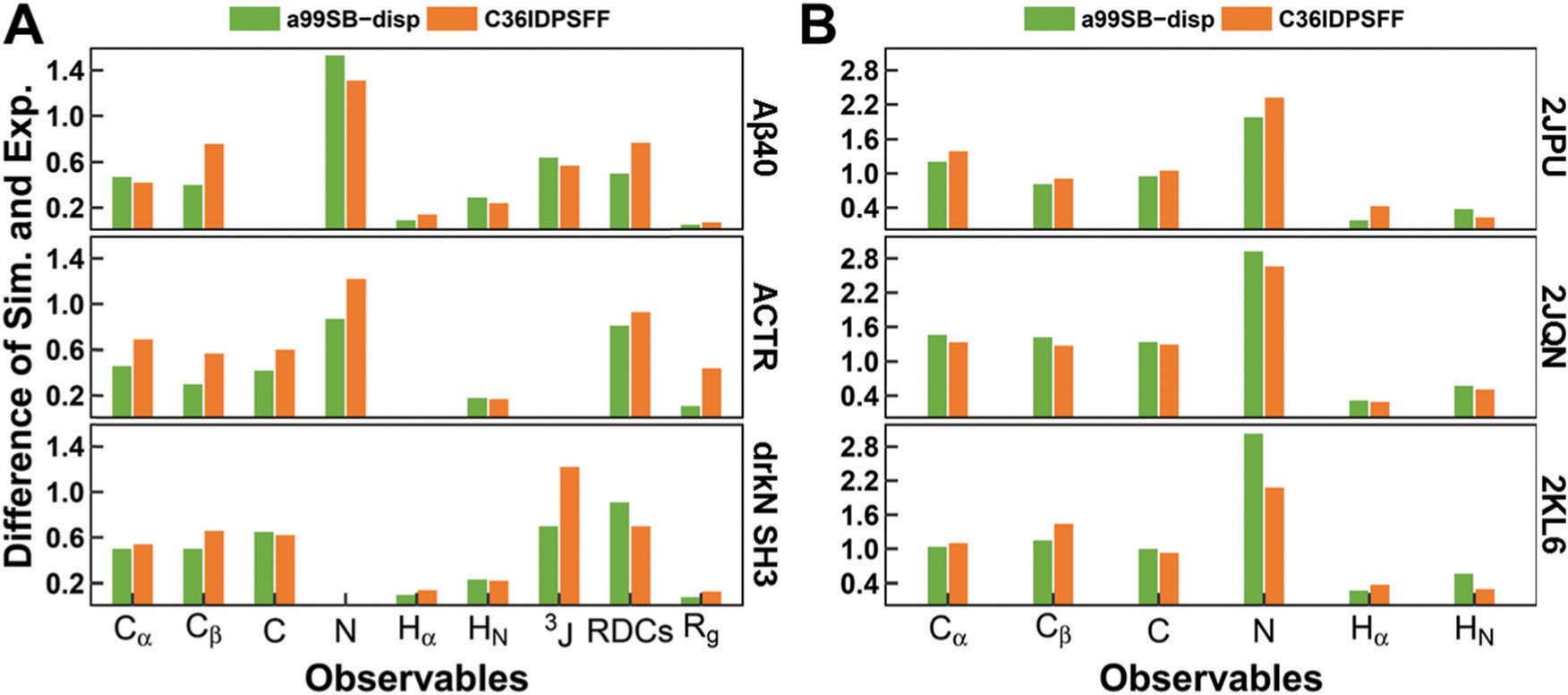
Difference between the experimental data and the simulated observables from C36IDPSFF in this study and a99SB-disp in previous work. (A) Disordered proteins, including Aβ40, ACTR, and drkN SH3. (B) Folded proteins, including 2JPU, 2JQN, and 2KL6. The differences of chemical shifts and J-couplings were estimated by RMS errors, and the differences of RDCs were estimated by Q factors. The differences of Rg were estimated by RgPenalty same as previous work. The 3JHNHa couplings were compared and referred to as 3J here. The missing values mean that the corresponding experimental data are not available.
