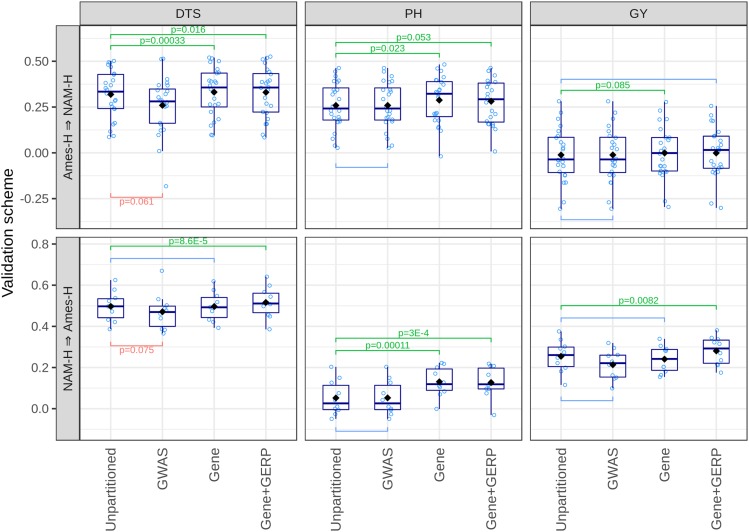
Figure 5.
Functional enrichment by gene proximity and GERP scores improves accuracy of genomic prediction models for DTS, PH, and GY. Prediction accuracy (y-axis): average correlation between observed and predicted genotype means in validation sets. Validation scheme: training panel ⇒ validation panel. Unpartitioned: DGBLUP (polygenic additive and dominance effects) in Ames-H, GBLUP (polygenic additive effects only) in NAM-H; GWAS, fixed effects at high-confidence QTL from GWAS; Gene, functional enrichment by proximity to genes (≤1 kb of an annotated gene); Gene+GERP, functional enrichments by proximity to genes and GERP scores. Black diamonds indicate average prediction accuracy. Significance of estimated differences in prediction accuracy (nonzero difference) was assessed by Student’s t-tests, paired by validation set. Only P-values < 0.1 (p) are shown. Ames-H, hybrid North Central Regional Plant Introduction Station association panel; NAM-H, hybrid U.S. Nested Association Mapping panel; GERP, genomic evolutionary rate profiling; GBLUP, genomic best linear unbiased prediction; DGBLUP, dominance GBLUP; DTS, days to silking; PH, plant height; GY, grain yield adjusted for DTS.