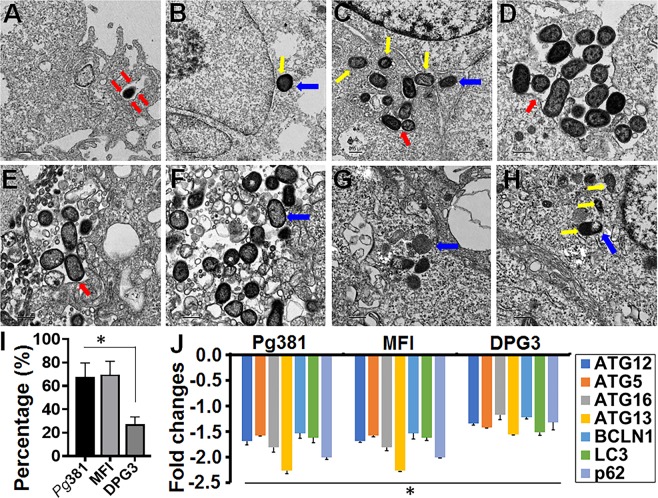
Figure 8.
P. gingivalis enclosed within single membrane structures or freely occupy the cytoplasm of ARPEs and escaped the autophagic vesicles. (A–H) Transmission electron microscopy (TEM) of single membrane structures within ARPEs infected with Pg381 (1 & 10 MOI), MFI and DPG3 (10 MOI) for 1 and 24 hours. All Pg strains shows mostly escaped the autophagic (double membrane) vesicles and enclosed within single membrane structures (red arrows) or freely occupy the cytoplasm (blue arrows). Yellow arrows show the bacteria in the cytosol around the nucleus. (A) Pg381 invaded into ARPEs enclosed within single membrane structure is evident at 1 MOI for 1 hour, whereas it reached the nucleus and lived-in the cytoplasm at 24 hours (B). As hypothesized, Pg381 (C,D), and MFI (E,F) were consistently detected within single membrane vesicles or freely occupy the cytoplasm of retinal epithelial cells, in contrast to our hypothesis that the DPG3 (G, H) also enclosed within the characteristics single membrane structures and apparently observed in the cytoplasm freely and close to the nucleus (yellow arrows) at 10 MOI for 24 hours. We also observed smaller numbers of DPG3 than the Pg381 and MFI both intra and extra cellularly (2 sets of representative images from each group). (I) The ratio of intracellular bacteria included in the single membrane were compared to total number of bacteria within ARPEs and plotted as percentage. Counting of the bacteria included in single membrane vesicles, freely occupy the cytoplasm or vacuoles after 24 hours of infection. Each strain was counted in six randomly selected grids for each sample. The ratio of DPG3 trapped in single membrane relative to the total intracellular bacteria was significantly lower than Pg381 and MFI. Refer the low magnification in Fig. S8 for the complete view. *P < 0.02. The analysis of the bacterial counts used Kruskal-Wallis test of different groups and Dunn’s test for multiple comparisons with 3 different experiments. (Scale bar:- A-H: 0.5 µm). J) TaqMan quantitative PCR results shows the genes related to autophagy machinery are significantly decreased by Pg381 (Mfa-1/FimA), MFI (FimA) and DPG3 (Mfa-1) infected ARPE-19 cells for 24 hours compared with uninfected control. This data is consistent with TEM imaging analysis (Figs. 5–8A–I) and confirm the lysosomal/vacuolar escape mechanism of P. gingivalis. The experiments were repeated thrice (biological) with three technical replicates (n = 9). Fold change in gene expression was normalized to the control and ≥ ±1.5 fold was considered as significant (*P < 0.05).