FIGURE 1.
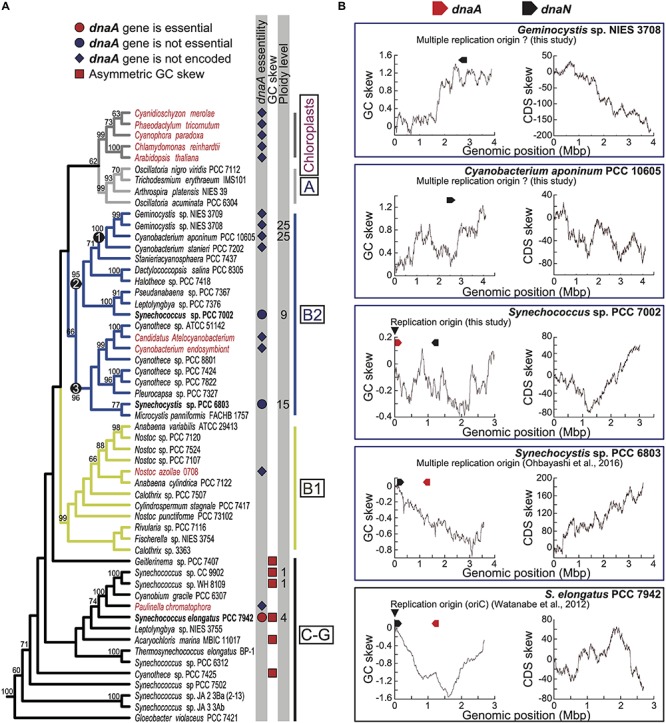
Phylogenetic distributions of dnaA among cyanobacterial and chloroplast genomes and GC and CDS skew profiles of chromosomes. (A) Phylogenetic relationships of cyanobacteria and chloroplasts and presence or absence of genomic dnaA. The tree was constructed using a maximum likelihood method based on 58 concatenated rDNA sequences (16S + 23S + 5S rDNA). Bootstrap values > 50% are shown above the selected branches. The full tree with the outgroup and accession numbers of respective nucleotide sequences are indicated in Supplementary Figure S1. Clades A, B1, B2, and C-G are defined according to a previous study (Shih et al., 2013). Chloroplasts and symbiotic species are shown in red, and free-living species are shown in black. The red or blue circle next to the species name indicates that dnaA was experimentally shown as essential or not essential for chromosome replication in the species (Ohbayashi et al., 2016; present study). The blue diamond next to the species name indicates the absence of dnaA. The red square next to the species name indicates that the chromosomal genome exhibits a clear asymmetric (V-shaped) GC skew profile. (B) Cumulative GC and CDS skew profiles of the chromosome of Geminocystis sp. NIES-3708, Cyanobacterium aponinum PCC 10605, Synechococcus sp. PCC 7002, Synechocystis sp. PCC 6803, and Synechococcus elongatus (profiles of other species are shown in Supplementary Figures S4, S5). The positions of dnaA and dnaN are indicated above the profiles. The arrowheads indicate the experimentally determined replication origins of Synechococcus sp. PCC 7002 (this study) and S. elongatus (Watanabe et al., 2012). The chromosome of Synechocystis sp. PCC 6803 is replicated from multiple origins (Ohbayashi et al., 2016).
