FIGURE 2.
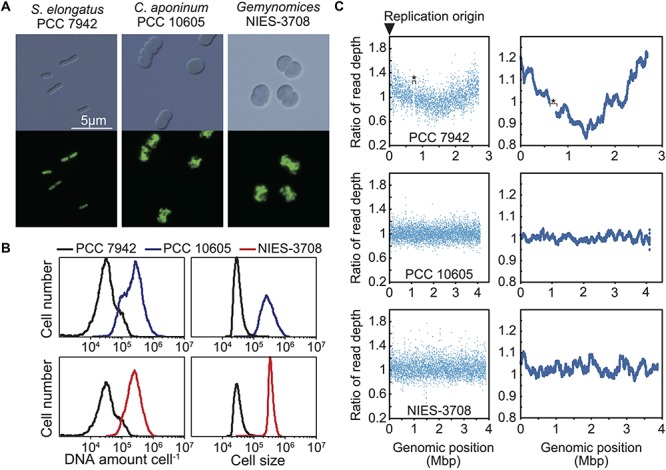
Ploidy and replication manner of the chromosomes of C. aponinum PCC 10605 and Geminocystis sp. NIES-3708. Exponentially growing C. aponinum (chromosome approximately 4.1 Mbp), Geminocystis sp. (chromosome approximately 3.9 Mbp), and S. elongatus (chromosome approximately 2.7 Mbp; 3–6 copies per cell; Zheng and O’Shea, 2017) (Supplementary Figure S2) were fixed and stained with SYBR Green and then examined using flow cytometry. (A) Images of SYBR Green-stained cells. Images were acquired using differential interference contrast microscopy (top) and fluorescence microscopy (bottom). (B) Distribution of DNA levels per cell and cell volumes of exponentially growing cultures of C. aponinum (blue), Geminocystis sp. (red), and S. elongatus (black). (C) Depths of the high-throughput genomic DNA sequence reads at their respective chromosomal regions. Genomic DNA was extracted from the exponentially growing cells (Supplementary Figure S2) and analyzed using an Illumina MiSeq System. Plots of 1-kb (left) and 100-kb windows (right). The number of reads (divided by the number of total reads) of the growing (replicating) cells normalized by that of the stationary phase (non-replicating) cells at each genomic position is shown. The asterisk in the profile of S. elongatus indicates a ∼50-kb genomic deletion in our wild type strain which has little effect on replication and cellular growth (Watanabe et al., 2012).
