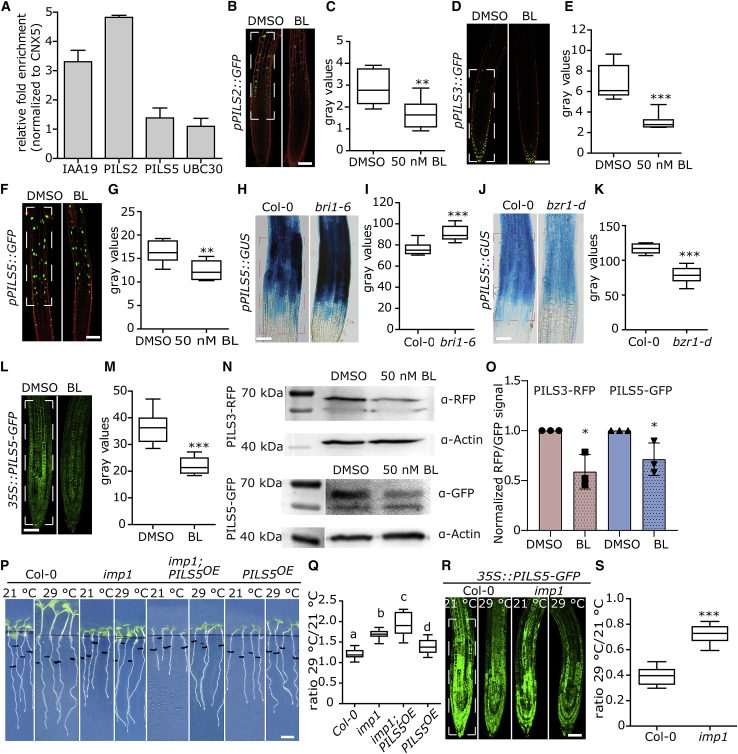
Figure 3.
BR Signaling Represses PILS Transcription and Protein Abundance
(A) Quantitative assessment of BZR1-CFP binding to promoters of PILS2 and PILS5, using chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR). Data represents means ± SD. Representative data of three replicates are shown. See also Figure S2. IAA19 and UBC30 are used as positive and negative controls, respectively. Data are normalized to negative control CNX5.
(B–G) Confocal microscopy images (B, D, and F) and quantifications (C, E, and G) of pPILS2::GFP (B and C, respectively), pPILS3::GFP (D and E, respectively), and pPILS5::GFP (F and G, respectively) expression patterns in roots treated with DMSO or 50 nM BL for 12 h (n = 8). Scale bar, 25 μm.
(H–K) GUS images (H and J) and measurements (I and K) of PILS5 promoter activity in main root of Col-0, bri1-6 (H and I, respectively), and bzr1-d (J and K, respectively). Scale bars, 25 μm.
(L and M) Confocal images (L) and quantification (M) of p35S::PILS5-GFP fluorescence after transfer on plates with DMSO or BL for 5 h, showing that BL reduces the PILS5 protein levels in roots of PILS5OE. Scale bar, 25 μm.
(N and O) Immunoblot with anti-RFP and anti-GFP antibody (N) and quantification (O) of signal intensity showing that BL downregulates PILS protein levels in p35S::PILS3-RFP and p35S::PILS5-GFP expressing seedlings. The α-actin antibody was used for normalization. The statistical evaluation shows the differences between the respective DMSO and BL application values. See also Figure S3.
(P and Q) Scanned images (P) and quantifications (ratio) (Q) of the root segment grown for 3 days at 21°C and subsequently transferred for another 3 days to 21°C (control) or 29°C (high temperature) (n > 20). Scale bar, 30 mm.
(R and S) Confocal images (R) and relative quantifications (S) of p35S::PILS5-GFP fluorescence in wild-type and in bri1 mutant (6 DAG) after exposure to 21°C (control) or 29°C (high temperature) for 3 h (n = 8). See also Figure S3. Scale bar, 25 μm.
h, hours; d, days. Stars and letters indicate values with statistically significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student’s t test in C, E, G, I, K, M, O, and S; p < 0.01, one-way ANOVA in Q). The dashed boxes represent the regions of interest (ROIs) used to quantify signal intensity.