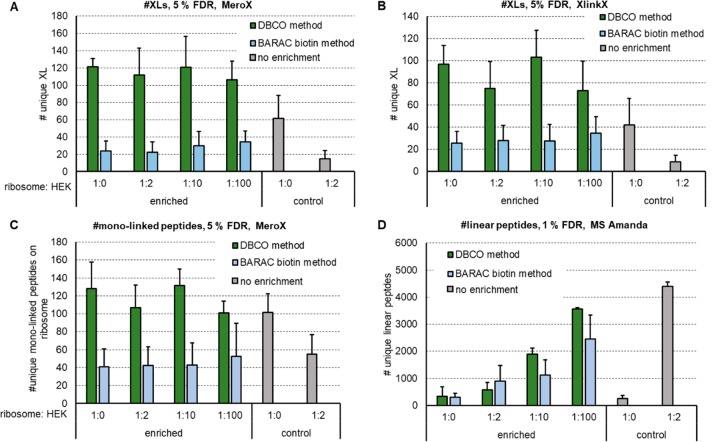
Figure 4.
Check for recovery of DBCO enrichment compared to the published BARAC biotin method by increasing a tryptic HEK background to mimic a cellular environment. Number of detected unique cross-links using MeroX (A) or XlinkX (B) for analysis, number of detected unique monolinked peptides via MeroX (C) and unique linear peptides via MS Amanda (D) after cross-linking 20 μg of E. coli ribosome each using 1 mM DSBSO. Tryptic HEK peptides were added to each sample in the given excess prior to enrichment with DBCO beads. Bars indicate the average values, with standard deviation depicted as error bar, FDR as indicated, n ≥ 3.