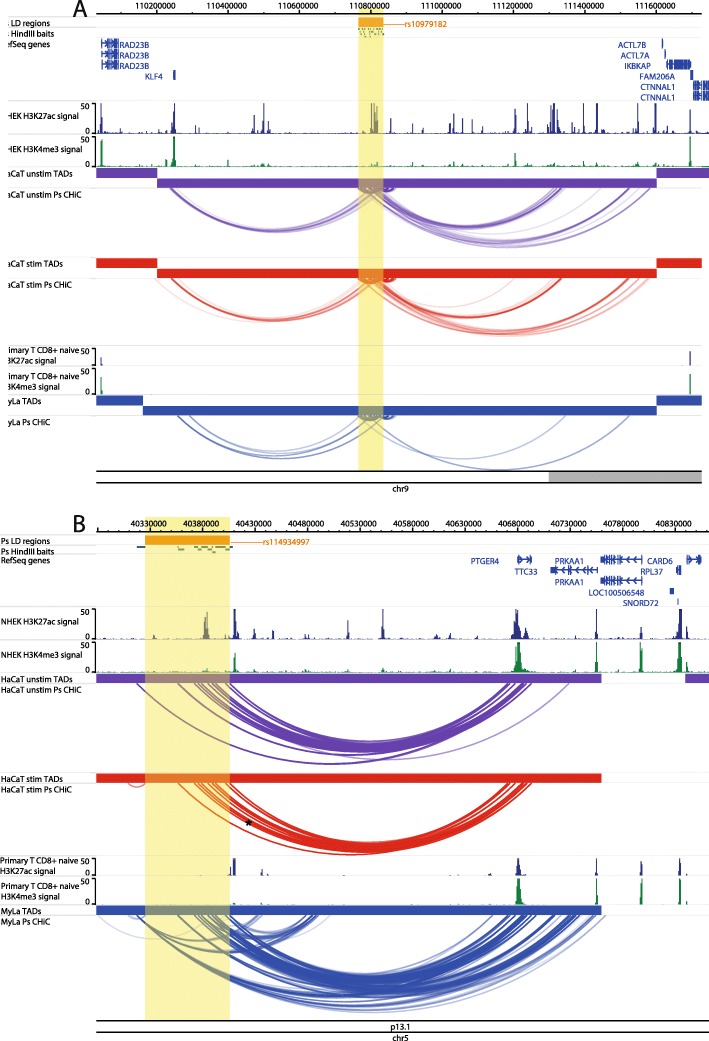
Fig. 1.
Examples of CHi-C Interactions implicating nearest/reported genes. Interactions are shown in the 9q31.2 (KLF4) locus (a) and the 5p13.1 (PTGER4) locus (b). The tracks include psoriasis (Ps) LD blocks as defined by SNPs in r2 > 0.8 with the index SNP, baited HindIII fragments, RefSeq genes (NCBI), H3K27ac and H3K4me3 P value signal in NHEK (ENCODE) and CD8+ primary naive T cells (Roadmap Epigenomics), TADs (shown as bars) and CHi-C interactions significant at CHiCAGO score ≥ 5 (shown as arcs) in three conditions: unstimulated HaCat cells (purple), HaCaT cells stimulated with IFN-γ (red) and My-La cells (blue). The highlighted region indicates the psoriasis LD block. The figure was made with the WashU Epigenome Browser, GRCh37/hg19 [31]