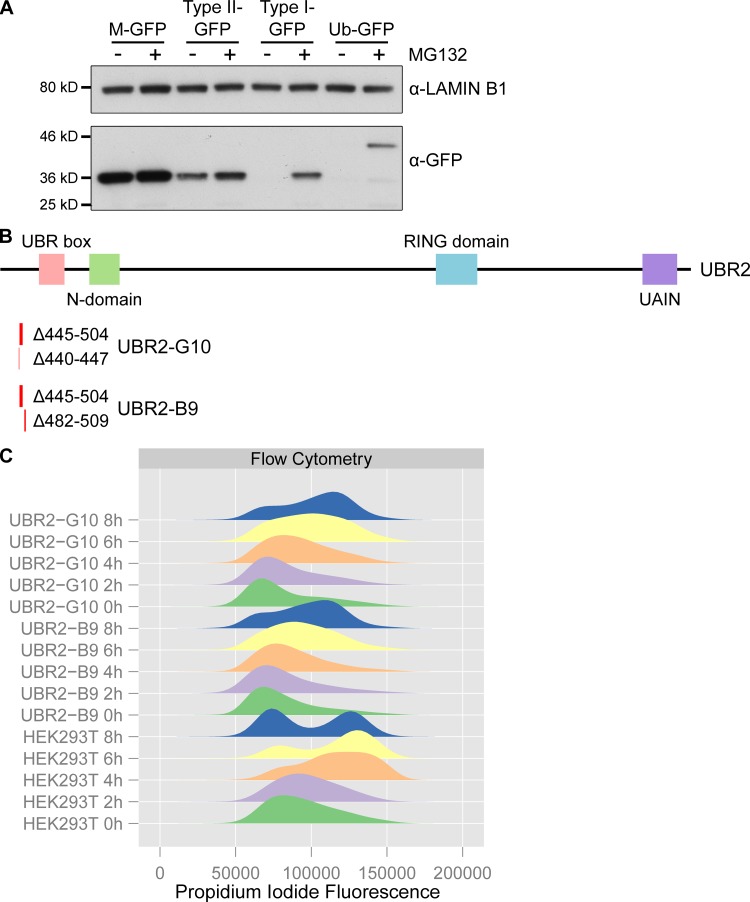
Figure S4.
Validation of N-end rule reporter and UBR2 mutant cell lines. (A) N-end rule GFP reporter cell lines are sensitive to proteasome inhibition. Ubiquitin fusion constructs that generate GFPs possessing N-end rule degrons (leucine for type II-GFP, arginine for type I-GFP) or methionine (M-GFP) at their N-termini, or a noncleavable Ub-GFP fusion construct, were stably integrated into Flp-In-293 cells. The abundance of the GFP reporters in these cell lines cultured in the presence or absence of the proteasome inhibitor MG132 was assessed by Western blotting using anti-GFP antibodies. Anti-lamin B1 antibodies were used as a loading control. (B) Location of UBR2 deletions recovered in compound heterozygous HEK293T clones generated by CRISPR/Cas9 genome editing. Locations and nucleotides deleted are indicated below a schematic showing the domain structure of UBR2 (Tasaki et al., 2005). Clone B9 has a 28-bp frameshift deletion (Δ482–509) in one UBR2 allele and a 60-bp in-frame deletion (Δ445–504) in another (numbering is relative to UBR2 cDNA NM_015255.2). Clone G10 has one allele with an 8-bp frameshift deletion (Δ440–447) in one UBR2 allele and the same 60 bp in-frame deletion (Δ445–504) present in clone B9 in the other. We did not recover any wild-type UBR2 sequences from these clones, or any CRISPR/Cas9-edited clones that only contained frameshift mutations in UBR2. (C) Clones B9 and G10 are slow growing and have altered cell cycle kinetics. Clones B9 and G10 exhibit cell death during a double thymidine block and release; therefore flow cytometry data represents a single thymidine block and release. Clones B9 and G10 have altered cell cycle distribution in the thymidine block (0 h) and are delayed in progression through to G2/M during the release by 2–4 h relative to the parental HEK293T cells.