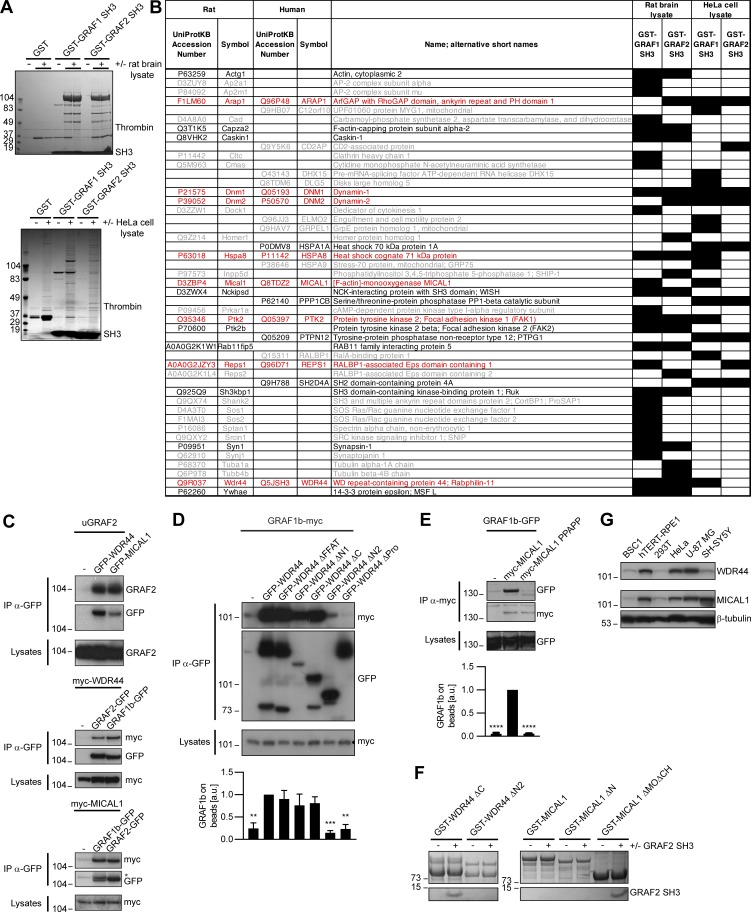
Figure S2.
GRAF1b/2 bind to MICAL1 and WDR44. (A) Purified GST, GST–GRAF1 SH3, or GST–GRAF2 SH3 was incubated with or without rat brain or HeLa cell lysates and pulled down on glutathione sepharose beads. Bound proteins were eluted by Thrombin digestion and visualized by electrophoresis and Coomassie staining. (B) Table summarizing proteins identified in (A) by LC-MS/MS. Cells filled in black indicate samples where the proteins were found. Proteins only identified in one pull-down are in gray (26/47 hits); those found both in GST–GRAF1 SH3 and in GST–GRAF2 SH3 pull-downs but only in one lysate are in black (13/47 hits); and those found both in rat brain and in HeLa cell lysates are in red (8/47 hits). Since GRAF1 and GRAF2 have a 67.9% identity and a 91.1% similarity in their SH3 domains, we focused our attention on these eight robust interacting partners. (C–E) Immunoprecipitation (IP) of transfected 293T cells with α-GFP (C and D) or α-myc–coated beads (E). (C) uGRAF2 was coimmunoprecipitated by GFP-tagged WDR44 and MICAL1. myc-tagged WDR44 and MICAL1 were coimmunoprecipitated by GFP-tagged GRAF1b and GRAF2. In the beads, GRAF2-GFP and GRAF1b-GFP overlapped with the remaining myc-MICAL1 band (*). (D and E) The efficiency of GRAF1b binding was quantified using coimmunoprecipitation with GFP-WDR44 (D) or myc-MICAL1 (E) as reference. n = 3–5. Data are means ± SEM; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Binding was abolished by removal (D) or mutation (E) of a proline-rich region. (D) Overexpressed WDR44 underwent proteolysis. (F) Pull-down test of GRAF2 SH3 by GST–WDR44 ΔC, GST–WDR44 ΔN2, GST-MICAL1, GST–MICAL1 ΔN, and GST–MICAL1 ΔMOΔCH. Beads were analyzed by electrophoresis and Coomassie staining showing direct binding of GRAF2 SH3 to WDR44 ΔC and to MICAL1 ΔMOΔCH. GRAF2 SH3 did not bind MICAL1 or MICAL1 ΔN. This suggests that the PPKPP motif of MICAL1 is not accessible in the isolated full-length protein and that in order to be exposed, MICAL1 has to undergo a conformational change, such as the one induced by Rab-binding (Schmidt et al., 2008). (G) Western blot analysis of an equal amount of BSC1, hTERT-RPE1, 293T, HeLa, U-87 MG, and SH-SY5Y cell lysates loaded on two replicate gels. One membrane was probed with α-WDR44, the other with α-MICAL1. β-tubulin was used as loading control.