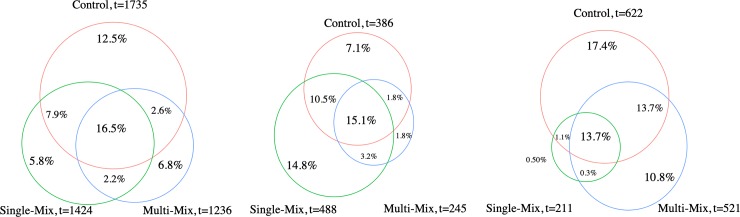
Fig 5. Change in unique bacterial types for each treatment at different time points.
All identified bacterial taxa were filtered by their respective ASV counts for individual samples. A. At time point 1, there were 1735 bacterial types from the control, 1424 from the single-mix and 1236 bacterial types from the multi-mix that had more than 1/3 of the replicates with nonzero ASV counts. Of all bacterial types analyzed, 16.5% had common bacterial type identifier codes, indicating the same type of bacteria, while unique bacterial types were identified for each treatment group. B. At time point 2, there were 386 bacterial types from the control, 488 from the single-mix and 245 bacterial types from the multi-mix that had more than 1/3 of the replicates with nonzero ASV counts. Of all bacterial types analyzed, 15.1% had common bacterial type identifier codes, indicating the same type of bacteria, while unique bacterial types were identified for each treatment group. C. At time point 3, there were 622 bacterial types from the control, 211 from the single-mix and 521 bacterial types from the multi-mix that had more than 1/3 of the replicates with nonzero ASV counts. Of all bacterial types analyzed, 13.7% had common bacterial type identifier codes, indicating the same type of bacteria, while unique bacterial types were identified for each treatment group.