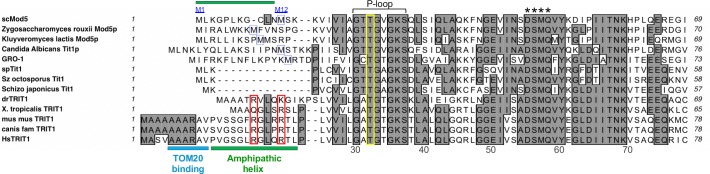
Fig 1. N-termini sequence alignment of representatives of phylogenetic groups of IPTases.
Sequences of the N-termini of IPTases. scMod5: Saccharomyces cerevisiae; GRO-1: IPTase from C. elegans; spTit1: Schizosaccharomyces (Sz) pombe; Sz octosporus; Sz japonicus; drTRIT1: D. rerio: Danio rerio; mus mus: Mus musculus; canis fam: Canis familiaris; X. tropicalis: Xenopus tropicalis; Hs: Homo sapiens. Numerals below the alignment show human amino acid numbering. The region between M1 and M12 of Mod5 that is required for mitochondrial targeting is indicated by a green rectangle above [34, 35]. The second methionines that may be used as alternative translation start sites for cytoplasmic localization are boxed in blue. The invariant Threonine within the conserved P-loop that functions in catalysis is in the yellow rectangle. Asterisks indicate the conserved DSMQ sequence that forms a network of contacts that mediate A37 recognition; the D of which acts as a general base for catalysis [24] and the M of which is M57 of TRIT1(57–467). As predicted by MitoFates [53], the TOM20-binding site of human TRIT1 is underlined with a blue bar and the amphipathic helix of TRIT1 and Mod5 are indicated with green bars. The two key basic residues in the conserved amphipathic helix of mammalian MTS that were mutated in human TRIT1 are in red rectangles.