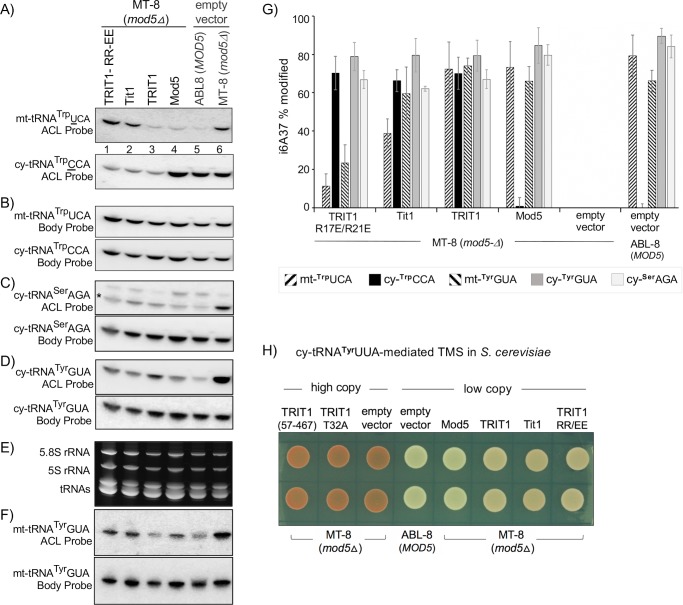
Fig 4. TRIT1 MTS-dependent modification of mt-tRNAs in S. cerevisiae.
A) A PHA6 i6A37-sensitive northern blot after transformation of the strains indicated above the lanes, showing sequential probing for mt-tRNATrpUCA and cy-tRNATrpCCA with the respective ACL-specific probes. MT-8 is mod5Δ and ABL-8 is a MOD5 positive control. B) The same blot showing reprobing (after stripping, Methods) with the body probes for mt-tRNATrpUCA and cy-tRNATrpCCA as indicated. C) The same blot showing ACL and body probings for cy-tRNASerAGA; asterisk indicates mature tRNA species, upper band is likely a cy-tRNASerAGA gene-specific precursor. D) The same blot showing ACL and body probings for cy-tRNATyrGUA with ACL and body probes as indicated. E) The small RNA region of the gel after staining with EtBr. F) Shows the ACL and body probe results for mt-tRNATyrGUA from a different blot. G) Quantitation of % i6A37 modification of the tRNAs in A-D, F from biological duplicate, or triplicate PHA6 blots as indicated; error bars indicate standard deviation (SD). H) Cy-tRNATyrUUA-mediated TMS in S. cerevisiae MT-8 (mod5Δ) transformed with empty vector or the IPTases indicated.