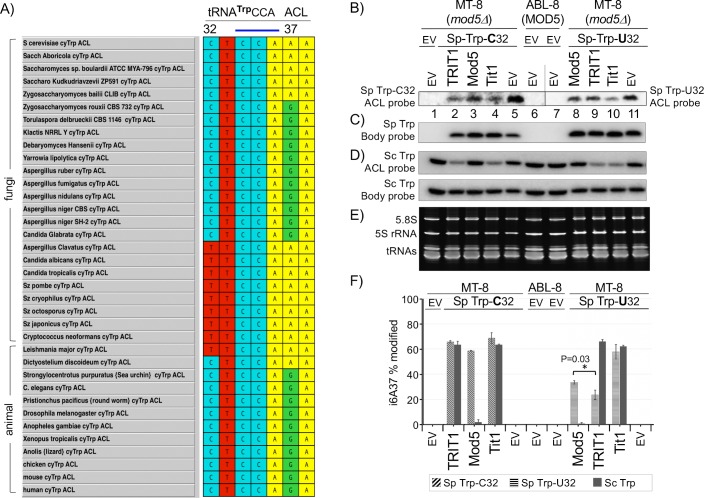
Fig 7. Position 32 as a determinant of differential IPTase activity for cy-tRNATrpCCA in eukaryotes.
A) Sequence alignment of the cy-tRNATrpCCA ACLs in the eukaryotes indicated; the 32 and 37 positions are numbered above; a horizontal blue bar indicates the AC. B) Results of PHA6 i6A37-sensitive PHA6 northern blot after expressing a S. pombe cy-tRNATrpCCA gene in MT-8 (mod5Δ) S. cerevisiae containing the native U32 (right panel) or this position mutated to C32 (left panel), or empty vector (EV) as indicated above the lanes, in the presence the IPTases or empty vector (EV) also indicated above the lanes; ABL-8 is a MOD5 positive strain. The right and left panel blots show separate hybridizations with ACL probes. Note that the order of the IPTases in the left and right panels differs, and carries through to panel F. C) The body probe result for S. pombe cy-tRNATrpCCA. D) ACL probe (upper) and body probe (lower) results for endogenous S. cerevisiae cy-tRNATrpCCA as indicated. E) shows the EtBr-stained gel from which the blot was made. F) Quantification of % i6A37 modification of the three different cy-tRNAsTrpCCA as indicated; error bars indicate standard deviation (SD) from biological duplicates. P value was obtained by the paired t-test.