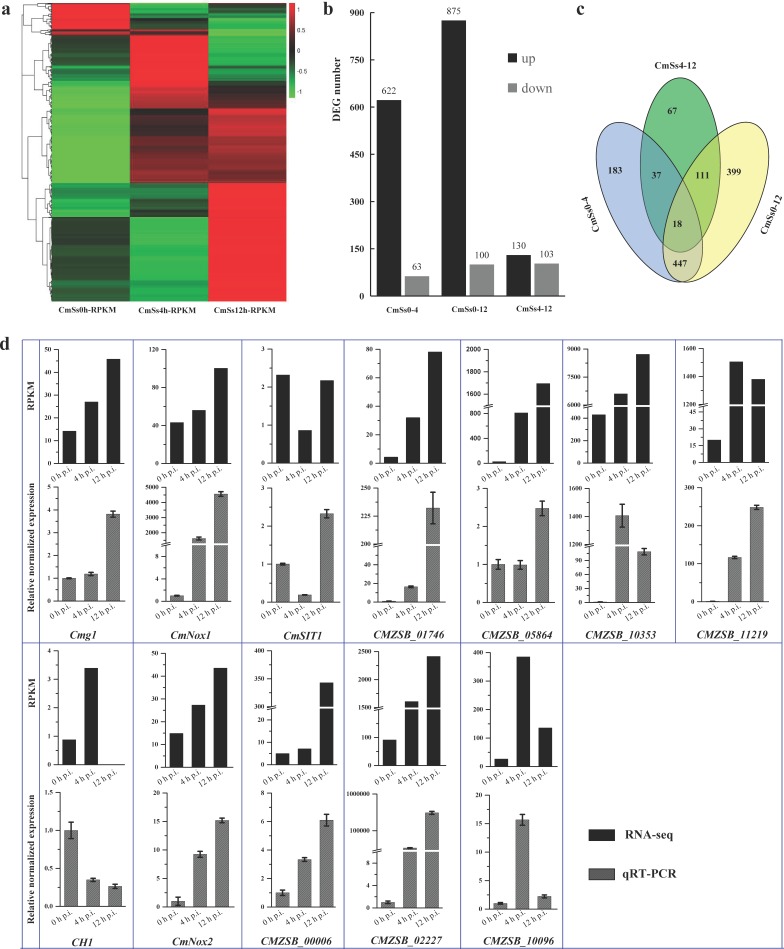
Fig. 2.
Statistical analysis of DEGs in C. minitans induced by hyphae of S. sclerotiorum and confirmation of the gene-expression pattern of the DEGs using qRT-PCR. (a) Based on the RPKM value, the heatmap of all the DEGs detected of C. minitans at the early stages with 0, 4 and 12 h induced by S. sclerotiorum. The detailed DEG list is presented in Table S3. (b) The statistical number of the up-regulated and down-regulated DEGs in the comparison pairwise of 4 h p.i. (CmSs0-4) and 12 h p.i. (CmSs0-12) to the control set of 0 h p.i. and 12 h p.i. to 4 h p.i. (CmSs4-12). (c) The common and particular DEGs in comparison groups of CmSs0-4, CmSs0-12 and CmSs4-12. (d) Confirmation of gene-expression pattern in C. minitans. Mycelial RNA of C. minitans parasitizing on S. sclerotiorum at 0, 4 or 12 h p.i. was used. The black bars represent the RPKM value of the genes in RNA-seq and the bars in grey shades show the qRT-PCR results of the DEGs. Error bars indicate the standard deviation of three replicates.