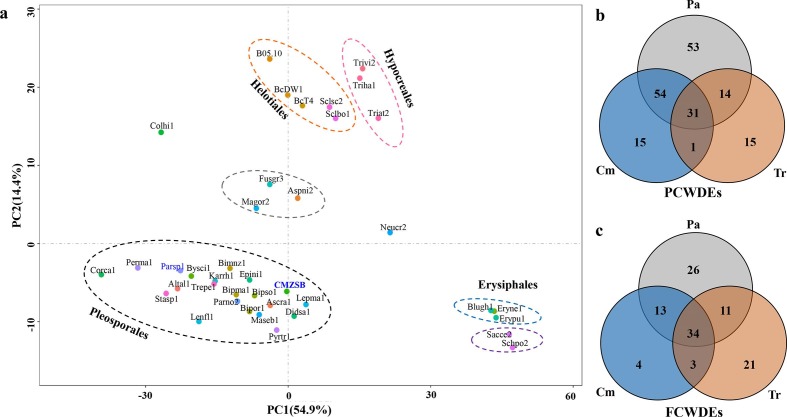
Fig. 3.
Comparison of CAZymes encoded in C. minitans ZS-1 and other 38 ascomycetes. (a) Multivariate grouping of ascomycete fungal genomes based on the protein number profiles of predicted CAZyme families using PCA. The CAZyme proteins from the fungal genomes were identified against the CAZy database. A number list of all CAZyme proteins in each genome is shown in Table S10. (b) Orthologous analysis of PCWDE-related protein comparison of C. minitans (Cm) with Trichoderma spp. (Tr) and seven plant pathogenic fungi (Pa); (c) PCWDE-related protein comparison of Cm with Tr and Pa. Orthologous groups were determined among the CAZymes encoded in the genomes of C. minitans, three Trichoderma spp. (T. atroviride, T. harzianum and T. virens) and seven plant pathogenic fungi (S. sclerotiorum strain 1980, S. borealis, B. cinerea strain DW1, B. cinerea strain B05.10, B. cinerea strain BcT4, P. nodorum strain SN15, and P. tritici-repentis strain Pt-1C-BFP) (details in Table S6). P(F)CWDEs, plant (fungus) cell-wall-degrading enzymes. Cm, CAZyme orthologous groups related to the cell wall in C. minitans; Tr, CAZyme groups related to cell wall encoded in Trichoderma spp.; Pa, CAZyme orthologous groups encoded by the seven plant pathogenic fungi genomes.