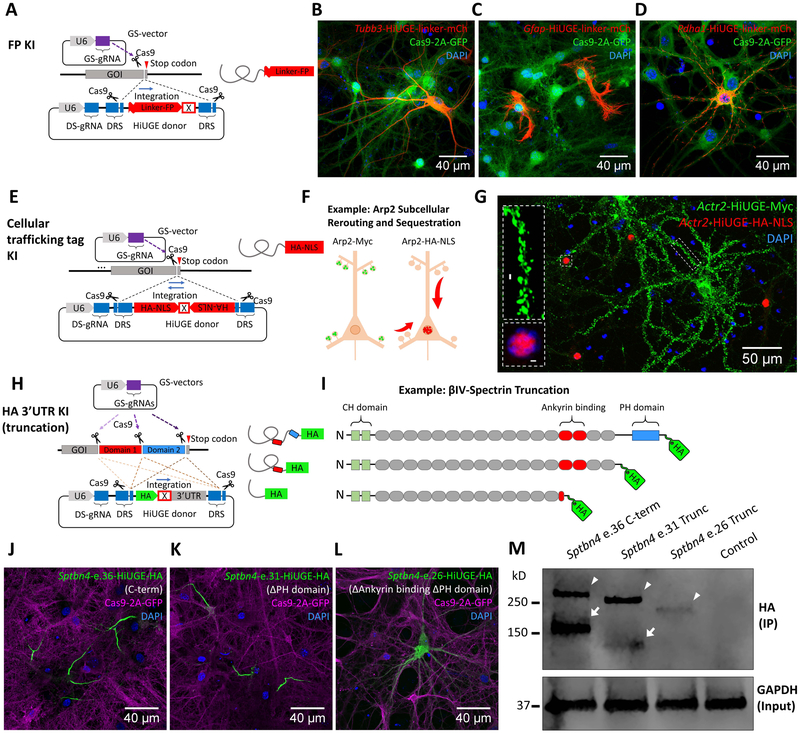
Figure 6. HiUGE Payloads Enable Flexible Selection Across Multiple Methods to Interrogate Endogenous Protein Functions.
(A) Schematic of fluorescent protein (FP) KI with flexible (GGGGS)4 linker. (B-D) Detection of mCherry (mCh) KI to different genomic targets (mouse Tubb3, Gfap, and Pdha1) with (B) antibody against mCh, or (C, D) direct visualization of mCh fluorescence. (E) Schematic of HiUGE subcellular re-routing constructs using cellular trafficking tags as the payloads. (F) Example using a HiUGE payload with HA-epitope and nuclear localization signal (HA-NLS) to sequester an actin cytoskeletal protein Arp2 to the nucleus. (G) Representative image of immunostaining following C-term HA-NLS KI to mouse Actr2 gene, showing the HA-NLS-tagged Arp2 (red) redirected to the nucleus. Simultaneously, the Myc-epitope (no NLS) tagged Arp2 (green) was enriched at the dendritic spines, consistent with the normal localization of Arp2. (H) Schematic illustration of HiUGE HA-3’ untranslated region (3’-UTR) KI to truncate endogenous proteins for conducting structure-function relationship studies. (I) Example using HA-3’UTR KI strategy to truncate ßIV-spectrin, composed of calponin-homology (CH) domains, spectrin repeats, and a pleckstrin homology (PH) domain. (J) C-term KI using a GS-gRNA targeting near the stop codon of the last coding exon 36 (e.36) of ßIV-spectrin is compared to (K) truncation of PH domain by targeting exon 31 (e.31), which still retains AIS enrichment, or (L) disruption of spectrin repeat 14 and truncation of downstream sequences by targeting exon 26 (e.26), which completely disrupts the AIS localization. (M) Western blot of HA-epitope showing stepwise reduction of molecular mass consistent with the serial truncation conditions (arrowheads). Arrowheads indicate the Σ1 isoform of ßIV-spectrin, while arrows indicate the Σ6 isoform. The Σ6 isoform of the truncation at exon 26 (e.26) appeared undetectable. Molecular mass is marked in reference to the ladder. Scale bar is indicated in each panel.