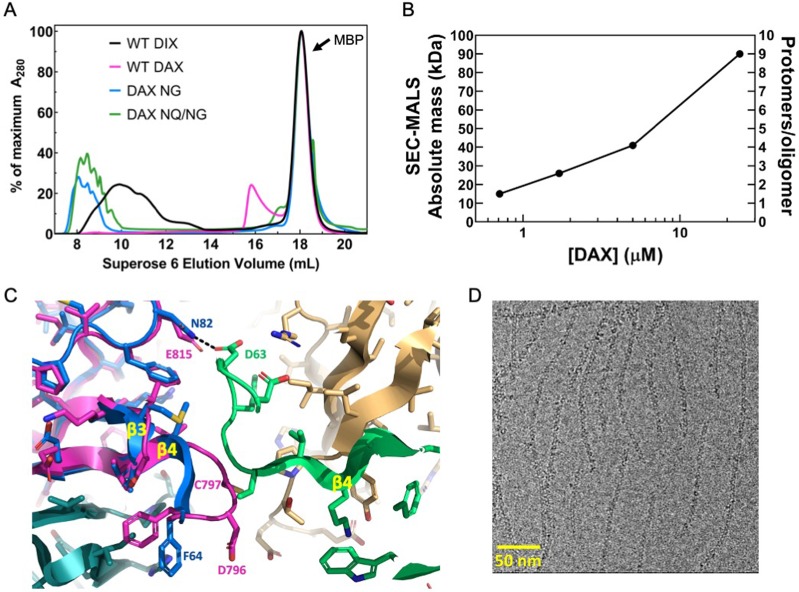
Figure 4. Oligomerization of Axin1 DAX and effects of oligomerization on signaling.
(A) Comparison of TEV-cleaved Axin1 DAX and Dvl2 DIX variants run on a Superose six size exclusion column (NG = E815N/E816G, NQ/NG = D793N/E794Q/E815N/E816G). Proteins were injected at 180 μM, and diluted to approximately 15 μM on the column. (B) SEC-MALS analysis of Axin1 DAX size as a function of concentration. (C) Superposition of the rat Axin1 crystal structure (PDB 1WSP; magenta) on one of the Dvl2 DIX domains (blue), showing potential clashes with another DIX protomer (green) across the inter-strand interface. Also shown is the substitution of N82 in DIX with E815 in DAX, which would eliminate the hydrogen bond with D63 of DIX and introduce electrostatic repulsion. Axin residue numbers are from the human Axin1 sequence. (D) Cryo-electron micrograph of DAX NQ/NG filaments.