Abstract
Clinical exome sequencing (CES) has become a routine diagnostic tool in several pediatric subspecialties, with a reported average diagnostic yield of ~25% in this patient poulation. The utility of CES in the United Arab Emirates (UAE) has not been previously investigated, most likely due to the lack of the appropriate tertiary pediatric centers and diagnostic genomic facilities in this country. Here, we report, for the first time, CES findings on a multispecialty pediatric cohort in the UAE (N = 51). This cohort, which was mostly Emirati (86%; 44/51), was followed at Al Jalila Children’s Hospital (AJCH), the first and only dedicated tertiary pediatric center in the country. CES demonstrates a high diagnostic yield (41%; 21/51) in this cohort, where 55% (28/51) had previous non-diagnostic genetic testing while for the remaining individuals (45%), CES was the first-tier test. Given the reported high consanguinity rate in this population, 48% of the positive cases (10/21) were due to genes associated with recessive conditions. However, 11 out of 21 positive cases (52%) were due to heterozygous pathogenic variants in genes known to cause dominantly inherited disorders, including a case with a dual diagnosis attributed to two different genes (2%; 1/51), and another case with a novel de novo variant and new phenotypic features for a known gene (2%; 1/51). Overall, we have identified 13 novel clinically significant variants and showed that application of CES as a first-tier test plays a significant role in genetic diagnosis and management of Emirati pediatric patients.
Abbreviations: DD, Developmental delay; ID, Intellectual disability; ASD, Autism Spectrum Disorders; MCA, Multiple Congenital Abnormalities
Keywords: Clinical exome sequencing, WES, Pediatric, Diagnostic yield
1. Introduction
Clinical exome sequencing (CES) is currently a routine pediatric diagnostic tool that has demonstrated an average diagnostic yield of 25% for patients with a wide range of indications [1], [2], [3]. The incidence of genetic disorders in the Arab countries, including the UAE, is extremely high and placing a burden of an estimated total annual cost of not less than $13 billion [4] to the economy. According to the March of Dimes report, the UAE is ranked sixth in the context of the prevalence of birth defects reaching up to 75.9 cases per 1000 live births [5]. Although the major known genetic illnesses include hemoglobin disorders (thalassemia, sickle cell), Down syndrome, and G6PD deficiency, the burden of other rare genetic disorders in the UAE has not yet been established partly due to lack of appropriate genetics diagnostic testing facilities [6]. The collective prevalence of those disorders is expected to be relatively high in this part of the world due to the high rate of consanguinity, extended family structure, and advanced paternal and/or maternal age at conception [7].
The utility of CES has been assessed in a small cohort from the UAE, though, this cohort was mainly focused on patients with highly suspected inborn errors of metabolism [8]. However, utility of CES across multiple other pediatric subspecialties remains to be explored. Al Jalila Children’s Specialty Hospital (AJCH) is the first dedicated tertiary pediatric center in the UAE. Therefore, its patient population presents with uniquely complex clinical scenarios across multiple specialties, of which genetics is most likely to be a major component. Since its opening in 2016, clinicians at Al Jalila have been utilizing CES for several indications as part of their clinical workup. This represents an unprecedented opportunity to explore the utility of CES in the UAE, and to start estimating the prevalence and types of rare genetic disorders in this country.
Here we demonstrate the high diagnostic yield of CES in a predominantly Emirati pediatric cohort presenting to multiple pediatric subspecialties at AJCH, and stratify this yield with respect to indication, age, gender, family history, and other demographics. We have also attempted to characterize the genetic landscape (i.e. genes and mutations) of the diseases for which a molecular diagnosis was made. Further, we detail the impact of CES diagnosis on medical management of patients.
2. Methods
2.1. Ethical approval
This study (AJCH-022) was approved by the Dubai HealthCare Authority (DHCA) Research Ethics Committee. Since this is a retrospective de-identified cohort description study, the ethics committee determined that an informed consent was not required.
2.2. Study design and WES testing
The period of this study was from July 2017 to January 2019, and the cohort included all patients seen as outpatient or inpatient at AJCH and for whom CES testing was ordered. There were no inclusion or exclusion criteria. This included 51 patients whose medical records were searched for the exome reports, indication, age, gender, nationality, and other lab or MRI/EEG imaging findings. CES during this period was performed at an external CAP-accredited clinical genetic testing laboratory. CES was performed using Agilent exome capture probes (Human All Exon V5 or V6) and Illumina sequencing platform at the testing facility where bioinformatics analysis and variant calling was done using a validated custom pipeline. Turnaround time for CES genetic diagnostic report release was approximately 12 weeks. We analyzed all reports from testing laboratory and defined “Diagnostic” reports as those that included variants classified as “pathogenic” or “likely pathogenic” according to the American College of Medical Genetics and Genomics (ACMG) guidelines for sequence variant interpretation [9] and that explained the patient’s underlying clinical condition. “Likely diagnostic” reports included rare, highly suspicious variants in genes matching patients’ clinical indications, though more information was needed at the time to confirm the pathogenicity of those variants according to the ACMG guidelines. The clinical utility of CES has been measured by evaluating medical charts to demonstrate the impact CES has on medical management and treatment strategies. The clinical factors measured include changes in patient care, medication or dietary changes, and further diagnostic testing.
3. Results
3.1. Cohort description
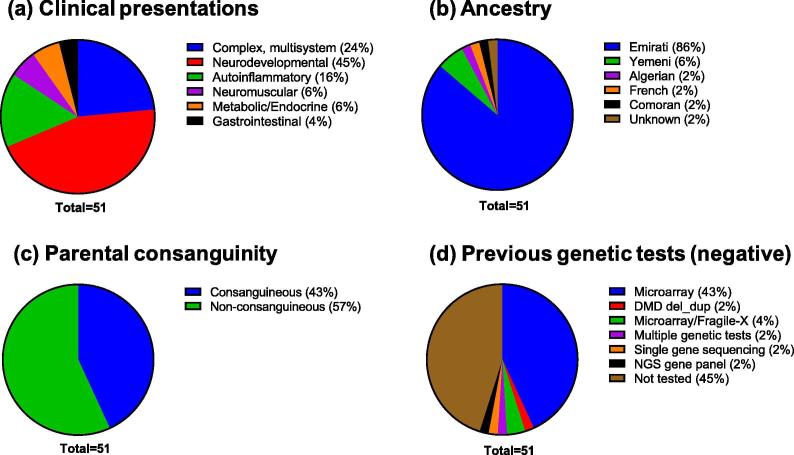
The studied cohort presented with diverse clinical indications typical of cases referred for CES. Based on the phenotypic information provided by the referring physician, the cases were classified into broad clinical categories. The largest category comprised of neurodevelopmental phenotypes (23/51, 45%), with global development delay (GDD), failure to thrive (FTT), hypotonia, seizures/epilepsy, intellectual disability and Autism Spectrum Disorders (ASD) as the most common indications (Fig. 1a). Patients with complex phenotypes involving multiple organ system abnormalities constituted 24% of the cases. Other categories were patients presenting with possible auto-inflammatory or rheumatology disorders, abnormalities of the neuromuscular system, abnormalities of the metabolic/endocrine system and gastrointestinal disorders (Fig. 1a).
Fig. 1.
Descriptive statistics of the patient cohort. The cohort included 51 consecutive cases referred for CES at AJCH (a) Primary disease classification of the patients based on clinical indications. (b) Ancestry of the cohort (c) Parental consanguinity in the cohort (d) Proportion of patients who had undergone other forms of genetic testing, prior to CES, which returned negative or normal results.
Of the 51 index cases, the largest proportion (44/51, 86%) were from the Emirati population (Fig. 1b). The age of the patients ranged from 2 months to 14 years with the largest group consisting of 1–5 years accounting to 43% (22/51). Approximately 10% of the patients were less than 1 year of age. Around 61% of the cohort were males. For 31 cases (61%), analysis of parents (trios) or an additional family member was performed, enabling investigation into possible modes of inheritance of disease in the family. Of note, 43% of the index cases were self reportedly from consanguineous families and 57% were from non-consanguineous families (Fig. 1c). In the consanguineous group, 63% were presented with positive family history while in the non-consanguineous group, 24% had positive family histories.
Prior to CES referral, 55% (28/51) of the patients had undergone extensive genetic diagnostic work-up, which were returned negative for the indication (Fig. 1d). The majority of the patients had been tested negative for chromosomal microarray analysis (CMA) alone (22/51, 43%) or in combination with other tests (~6%). Other tests included karyotyping, single-gene testing, next-generation sequencing (NGS) gene panels and Duchenne Muscular Dystrophy (DMD) gene deletion/duplication analysis (Fig. 1d). One patient had been tested negative for 4 tests including karyotype, CMA, methylation analysis, and an NGS gene panel.
3.2. Diagnostic yield of CES
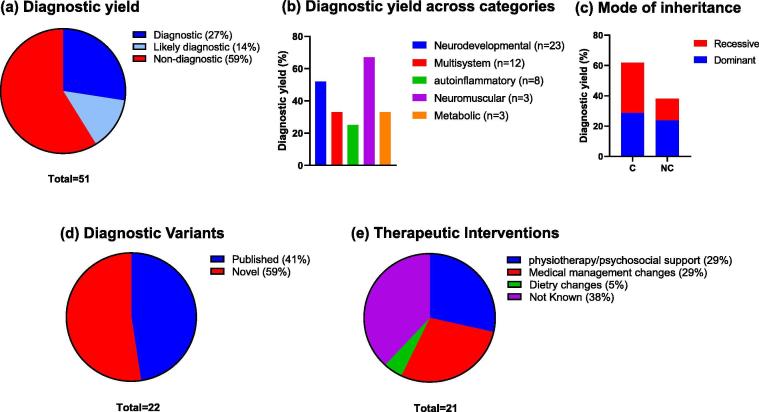
Overall, 41% (21/51) CES referrals harbored one or more variants that satisfied the criteria for a full or partial genetic diagnosis (Fig. 2a). The diagnostic yield across different clinical categories are shown in Fig. 2b. Among the major clinical categories, the highest diagnostic rate was observed in the neuromuscular disease category (67% or 2/3) followed by neurodevelopmental category (52% or 12/23). Despite the high rate of consanguinity, variants identified for 11 out of the 21 cases were heterozygous for dominant disorders (Fig. 2c). When we analyzed the data for consanguineous and non-consanguineous groups separately, the consanguineous group was found to have a higher number of recessive homozygous variants (n = 7) than the non-consanguineous group (n = 3) (Fig. 2c). Parental consanguinity was observed to be linked to a higher diagnostic rate since the observed diagnostic yield for consanguineous group was 59% (13/22) while that for the non-consanguineous group was 28% (8/29). Among CES positive cases 11 out of 21 had undergone previous genetic tests that were unrevealing.
Fig. 2.
Descriptive statistics of the positive exome cases. A total of 21 index cases out of 51 referred for WES received a positive diagnosis. (a) Overall diagnostic yield of CES across all disease categories (b) Diagnostic yield of CES in specific clinical categories. (c) Proportion of dominant and recessive disorders among consanguineous versus non-consanguineous positive cases. “C” indicates consanguineous, "NC" indicates non-consanguineous. Number of total identified variants were 22 across 21 positive cases. (d) Proportion of novel variants identified in the cohort. Novel variants are defined as variants that were absent from population databases such as gnomAD and dbSNP, and disease databases such as ClinVar and HGMD and published literature (refer to Suppl Table S1) (d) Proportion of diagnostic/likely diagnostic cases receiving an altered management plan as indicated.
The diagnostic or likely diagnostic cases included a diverse array of disorders such as infantile hypotonia, with psychomotor retardation and characteristic facies 1; (IHPRF1; NALCN), Aspartylglucosaminuria (AGA), Epileptic encephalopathy, early infantile, 4 (STXBP1) and severe myoclonic epilepsy (SCN1A). In patients with complex phenotypes the molecular diagnoses included Ehlers-Danlos syndrome, kyphoscoliotic type, 1 (PLOD1), Bardet-Biedl syndrome 7 (BBS7) and Weaver syndrome (EZH2) suggesting highly heterogeneous syndromes can be resolved by CES. One patient in this category presenting with GDD, motor delay and infections received dual diagnosis for two autosomal dominant disorders attributed to the presence of heterozygous pathogenic variants in two different genes (KMT2C; OMIM#617768 and GNAO1; OMIM #617493) associated with those disorders (Table 1). The spectrum of variants included missense (n = 9), nonsense (n = 7), frameshift (n = 4) and splice-site (n = 1) variants. One case was heterozygous for a deletion at the chromosomal locus 9q34.3. About 59% of the variants identified by CES were previously unpublished in the peer-reviewed literature or disease variant databases (Fig. 2d) which included 7 novel loss-of-function variants and 6 novel missense variants (Table 1, Supplementary Table S1).
Table 1.
Diagnostic and likely diagnostic CES cases with clinical indications and details of variants identified.
| ID | Age | Gender | Ancestry | Case | Family History | Parental Consanguinity | Primary Disease classification | Genes | Variants (cDNA; protein) | Variant class/ status | Mode of Inheritance | Zygosity | Therapeutic interventions |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 Yrs. | M | Emirati | Cerebro-oculo-facio-skeletal (COFS) syndrome, GDD. | Positive in first degree cousins | Yes | Neurodevelopmental | ERCC5 | NM_000123.3: c.2902 T > C; p.Trp968Arg | VUS/ Novel# | AR | Hom. | Occupational therapy, physiotherapy, Postponed surgical procedure |
| 2 | 14 Yrs. | M | Emirati | GDD, FTT, RF, Cerebral palsy, generalized tonic colonic seizure | Mother: Hyperactive thyroid, undergone thyroidectomy, currently on thyroid replacement therapy, brother: developmental delay & hypothyroidism, maternal uncle: muscle weakness & wheelchair bound | No | Neurodevelopmental | NALCN | NM_001350748.1:c.2621_2622insATACTAAA; p.Phe875Tyrfs*2 | Pathogenic/Novel | AR | Hom. | Intermittent non-invasive BIPAP, occupational therapy, physiotherapy, dietary management |
| 3 | 19 Mos. | M | Emirati | Rubinstein taybi syndrome, Recurrent seizures, GDD, FTT | Sister: Congenital hypothyroidism, 2 maternal aunts: hypothyroidism | Yes | Neurodevelopmental | TSHR | NM_000369.2: c.1295A > G; p.Asn432Ser | VUS/Novel# | AR/AD | Hom. | Neuro rehabilitation, physiotherapy, speech therapy |
| 4 | 10 Yrs. | F | Emirati | GDD secondary to anoxic brain injury, intellectual disability | None | Yes | Neurodevelopmental | TRAPPC6B | NM_001079537.1: c.267 + 1G > A; p.? | Pathogenic/Novel | AR | Hom. | School for special needs, occupational therapy, psychology care |
| 5 | 7 Yrs. | M | Emirati | Bardet-Biedl Syndrome, Epilepsy | None | Yes | Complex, multisystem | BBS7 | NM_176824.2: c.124G > A; p.Gly42Arg | VUS/ Novel# | AR | Hom. | Multiple specialties follow up |
| 6 | 3 Yrs. | M | Emirati | ASD, G6PD, GDD | Mother: G6PD, One maternal cousin: delayed walking | Yes | Neurodevelopmental | TSC2 | NM_000548.4: c.1754G > A; p.Arg585His | VUS/ Reported | AD | Het | Speech therapy, occupational therapy |
| G6PD┼ | NM_000402.4: c.653C > T; p.Ser218Phe | Pathogenic/Reported | XLD | Hemi | |||||||||
| 7 | 5 Yrs. | M | Emirati | Central DI, CVID, ICF, GDD, FTT, OSA | Paternal cousin: cleft lip, mother: DMT1 | Yes | Endocrine/Metabolic | BCL11A | NM_022893.3: c.317C > T; p.Thr106Met | VUS/ Novel | AD | Het. | Monitoring of fluid intake, low salt diet, Life-style changes, caregiver escort |
| 8 | 3 Yrs. | M | Emirati | Acute gastritis, Epilepsy | Brother: 4 years old, speech delay. Father: Delayed speech when he was young | No | Neurodevelopmental | FGFR3 | NM_001163213.1: c.749C > G; p.Pro250Arg | Pathogenic/Reported | AD | Het. | Information unavailable |
| 9 | 7 Mos. | F | Comoros | Clinical Vasculitis Suspecting Auto inflammatory disease, Noma Neonatorum, SCID | Parental aunt and baby's father with similar lip condition | No | Autoinflammatory | deletion chromosome 9q34.3 (Kleefstra Syndrome) | Pathogenic/Known syndrome | AD | Het. | Follow-up with sub-specialty (Infectious disease) | |
| 10 | 13 Mos. | M | Emirati | Left upper extremity shaking, Channelopathy | Maternal first cousin: 13 year old girl having epilepsy and on AED Paternal second cousin: Born with low oxygen at birth and has epilepsy |
Yes | Neurodevelopmental | SCN1A | NM_001165963.2: c.5536_5539del; p.Lys1846Serfs*11 | Pathogenic/Reported | AD | Het. | Information unavailable |
| 11 | 11 Yrs. | F | Emirati | ADHD | Epilepsy in fathers cousin | Yes | Neurodevelopmental | STXBP1 | NM_003165.3: c.1099C > T; p.Arg367* | Pathogenic/Reported | AD | Het. | Information unavailable |
| 12 | 5 Yrs. | M | Emirati | ASD, GDD | Second of 3 children. 2 Siblings: Similarly affected. 7 yrs old girl (mild symptoms) and 3 yrs old boy (Severely affected) Mother has Intellectual disabilities. Father with borderline IQ Uncle: 24yrs, father's brother, with ASD. |
No | Neurodevelopmental | CIC | NM_015125.4: c.1927G > C; p.Gly643Arg | VUS/ Novel | AD | Het. | Information unavailable |
| 13 | 8 Yrs. | M | Emirati | DD, Epilepsy, ASD, Hyperactivity | None | No | Neurodevelopmental | SHANK3 | NM_033517.1: c.4984C > T; p.Gln1662* | Likely pathogenic/Novel | AD | Het. | Information unavailable |
| 14 | 10 Mos. | F | Emirati | Heart murmur, congenital joint contractures | None | No | Complex, multisystem | PLOD1 | NM_000302.4: c.955C > T; p.Arg319* | Pathogenic/Reported | AR | Hom. | Information unavailable |
| 15 | 15 Mos. | F | Emirati | Complex febrile seizure, DD | Diabetes: Grandparent. | Yes | Neurodevelopmental | WDR45 | NM_007075.3: c.19C > T; p.Arg7* | Pathogenic/Reported | XLD | Het. | Information unavailable |
| 16 | 2 Mos. | M | Emirati | Seizure, Dysmorphic features, Infection | Mother: alpha thalassemia carrier, father healthy, maternal grandmother had multiple abortions unknown reason. First child died at 5 years of age diagnoses: Mother had 3 other miscarriages early trimester, not investigated |
No | Complex, multisystem | EZH2 | NM_004456.4: c.44G > T; p.Trp15Leu | VUS/ Novel | AD | Het. | Intubation, seizure medications, physiotherapy, occupational therapy, speech therapy, Respiratory therapy, diet monitoring. |
| 17 | 10 Mos. | F | Emirati | URTI, DD, Dysmorphic Features | None | Yes | Complex, multisystem | KMT2C | NM_170606.3: c.9391C > T; p.Gln3131* | Likely pathogenic/Novel | AD | Het. | Medication changes |
| GNAO1 | NM_020988.2: c.680C > T; p.Ala227Val | Pathogenic/Reported | AD | Het. | |||||||||
| 18 | 12 Yrs. | M | Emirati | Muscular Weakness, myopathy, bilateral calf hypertrophy | None | Yes | Neuromuscular | SGCA | NM_000023.2: c.292C > T; p.Arg98Cys | Likely Pathogenic/Reported | AR | Hom. | Physiotherapy |
| 19 | 10 Yrs. | M | American (origin Yemen) | Sever progressive arthropathy, short trunk dysplasia with suspicion of pseudo-rheumatoid dysplasia | Mother had one miscarriage and one baby died after birth (forceps delivery), one sister with similar features, other sister and brother normal. They have a sister that has 6 offspring, four of them have crippling arthropathy that progress to disability for unknown etiology | Yes | Autoinflammatory | WISP3 | NM_003880.3: c.707delG; p.S236Tfs*5 | Pathogenic/Novel | AR | Hom. | Information unavailable |
| MEFV┼ | NM_000243.2: c.2230 G > T; p.A744S | Pathogenic/Reported | AR | Het. | |||||||||
| 20 | 6 Yrs. | M | Emirati | expressive speech delay, paraplegia, gait disturbance, DD | None | No | Neuromuscular | KMT2B | NM_014727.2: c.3043C > T; p.Arg1015* | Pathogenic/Novel | AD | Het | Rehabilitation, speech therapy, Specialty follow-up |
| 21 | 6 Yrs. | M | Emirati | GDD, ASD | Paternal uncle has epilepsy, 4 years old brother has speech delay but walked on time | Yes | Neurodevelopmental | AGA | NM_000027.3: c.199G > T; p.Glu67* | Likely pathogenic/Novel | AR | Hom. | Enrolled in special needs center, continued follow- up |
#Novel nucleotide change causing different amino acid substitution at a previously reported codon position; ┼ Additional findings related to the phenotype reported prior to CES and confirmed by CES.
The Abbreviations: Yrs.: years Mos.: months M: male F: female DD: developmental delay GDD: global developmental delay FTT: failure to thrive RF: respiratory failure G6PD: glucose-6-phosphate dehydrogenase ASD: Autistic Spectrum Disorder DI: diabetes insipidus CVID: common variable immunodeficiency disease OSA: obstructive sleep apnea ICF: immunodeficiency-centromeric instability-facial anomalies syndrome SCID: Severe combined immunodeficiency ADHD: Attention Deficit Hyperactivity Disorder URTI: upper respiratory tract infection AD: Autosomal dominant AR: autosomal recessive XLD: X-linked dominant Het.: Heterozygous Hom.: Homozygous Hemi: Hemizygous.
Out of the 21 cases with a genetic diagnosis, 14 had a definitive molecular diagnosis with the identification of pathogenic or a likely pathogenic variant (Fig. 2a, Table 1). Variants of unknown significance (VUS), which we still considered as “likely diagnostic”, were reported in seven cases (Fig. 2a). These variants were included in the list of positive cases because they were rare variants with strong evidence of pathogenicity (segregation, different amino acid changes at the same position, etc.), and were in genes highly matching the patient’s reported phenotype and the inheritance model. For example, the heterozygous c.1927G > C (p. Gly643Arg) variant of uncertain clinical significance, detected in the CIC gene, was identified in three affected siblings from the same family. Pathogenic variants in the CIC gene cause mental retardation, autosomal dominant 45 (OMIM#617600), characterized by variable intellectual disability, often with language delay and attention deficit/hyperactivity disorder (ADHD), and autism spectrum disorder [10]. Parental analysis was also included in this study, which revealed that this variant was inherited from the father, with borderline IQ. Another selected example involves a novel missense variant in BCL11A that occurred de novo in a patient with suspected metabolic/endocrine syndrome and dysmorphic features that was able to explain the patient phenotype partially. Heterozygous de novo mutations in the BCL11A gene are associated with Diaz-Logan syndrome characterized by intellectual disability, variable dysmorphic features and asymptomatic persistence of fetal hemoglobin [11]. This patient had additional renal phenotypic features that are not currently associated with the variations in the BCL11A gene. However, a contiguous gene deletion syndrome involving the BCL11A and adjacent genes in the 2p15p16.1 interval is associated with renal and cardiac anomalies in addition to brain anomalies, though those phenotypes are largely attributed to other genes [12], [13]. Emerging information on the genes may help to reclassify these variants in the future and provide better clinical correlation.
3.3. Impact of CES on medical management
Importantly, several of the positive diagnoses had implications for the patients and their families. Altered medical management was reported in 13 out of 21 patients (62%). Physiotherapy, occupational therapy, speech therapy and other forms of psychosocial supports were initiated for 6 patients (Fig. 2e). Although such therapies could have been started anyways, genetic diagnosis reinforced those management therapies and ruled out any possible underlying serious disorders for which other management plans would have been more appropriate. Medical management changes such as altered medications, treatment plan or specialties follow-up were reported for another 6 patients (Fig. 2e). Dietary changes were implemented for one patient. An example is case No.2 presenting with global developmental delay, generalized muscle wasting, crowded teeth, scoliosis, dysmorphic features, epilepsy, and mild hypotonia, who received a positive diagnosis for infantile hypotonia, with psychomotor retardation and characteristic facies 1 (IHPRF1) related to NALCN gene. Although no specific treatment plan was available for this disorder, the CES findings excluded other possible differential diagnoses and supported a management plan including intermittent non-invasive bilevel positive airway pressure (BIPAP), occupational therapy, physiotherapy, and dietary management.
4. Discussion
We have reported here the overall diagnostic yield of 41% for CES in a complex pediatric cohort mainly comprised of Emirati patients. This yield is higher than the reported average CES yield of 25% [2], which can vary based on indication reaching up to ~50% for hearing and vision disorders [14], [15]. Although larger cohort sizes are needed for more generalizable conclusions, our findings from this small cohort (n = 51) did provide interesting insights into the incorporation of whole exome sequencing into clinical practice and affirm previous observations about the influence of cultural practices and population structure on genetic disorders. The cohort presented here had diverse and striking phenotypes that belonged to different clinical categories. The highest diagnostic yield (52%) was obtained in the neurodevelopmental category which encompassed patients presenting with more than one phenotype related to developmental delays, seizures, hypotonia and dysmorphisms emphasizing the fact that CES can resolve many underlying genetic conditions in this category. Since AJCH is a tertiary pediatric care hospital, most patients selected for CES had undergone extensive clinical and diagnostic work-up, which would have helped the physicians to eliminate other underlying reasons making a positive CES result highly probable. At present, for individuals with unexplained DD/ID, ASD, or MCA, chromosomal microarrays (CMA) are recommended as the first-tier test with a reported diagnostic yield of 15%–20% [16]. A recently reported meta-analysis led to a consensus statement that among patients with neurodevelopmental disorders, whole exome sequencing is likely to produce a higher molecular diagnostic yield than CMA and should be considered as a first-tier test [17]. Among the 21 CES positive patients in our cohort, 10 had been previously tested negative in CMA analysis indicating the trend of higher diagnostic utility of CES.
The prevalence of genetic disorders is attributed to the peculiar demographic features of UAE which is characterized by large family size, advanced maternal and paternal age and consanguinity in the ranges of 25–60% [7]. In our cohort comprising mostly of Emirati individuals (86%), around 43% were from consanguineous unions (Fig. 1c). Among the hereditary disorders, recessively inherited disorders appear to be predominant owing to the high rates of consanguinity [4], [7]. Surprisingly, in our cohort dominant disorders were found to be more common than recessive disorders. However, when we analyzed the consanguineous group and non-consanguineous group separately, autosomal recessive disorders were more common in the consanguineous group than in the non-consanguineous group. The consanguineous group had a higher diagnostic yield than the non-consanguineous group with recessive and dominant disorders in equal proportions (Fig. 2c). Some variants in the autosomal dominant category were de novo or presumed de novo. Paternal age-related de novo mutations have been reported to confer high risk for autism spectrum disorder (ASD), congenital heart disease, neurodevelopmental disorders with epilepsy, intellectual disability and schizophrenia (SCZ) [18], which requires additional investigations in this cohort.
Large scale studies on genetic variations in this unique population are lacking and could prove to be a bottleneck for identifying and classifying rare variants since this population is underrepresented in the popular public databases such as gnomAD. The Greater Middle East (GME) Variome Project [19] catalogues coding base variations from 2,497 individuals in the greater Middle East region, namely the Arabic Peninsula, Northeast Africa, Syrian Desert, Central Asia, Northwest Africa and Turkish Peninsula. Although very useful, this study is very small and does not represent the full normal variation in all subpopulations within this highly diverse region.
Variants that are rare in other public databases, such as gnomAD, could be common in this population and can confound exome analysis. Among the positive variants identified, 59% were novel variants not published before, either in the literature or public databases, revealing population-specific disease variants. Some of the disorders uncovered through CES were found to be disorders having a higher prevalence in the Arab population or clustering to Arab communities or isolates. For example, Ehlers-Danlos syndrome caused by the PLOD1 gene has been reported to cluster in Bedouin families in UAE [20]. Bardet-Biedel syndrome is reported to be highly prevalent in Arab populations [20]. Comorbidity is another factor contributing to diverse clinical presentations patients from the region especially with major disorders including thalassemias, sickle cell disease, and cystic fibrosis [4]. Patient No.6 was diagnosed with G6PD deficiency and the occurrence of a G6PD pathogenic variant had been earlier confirmed. In this patient a VUS in the TSC2 gene was identified which could partially explain their ASD/GDD features. Another patient received a dual diagnoses for two different autosomal dominant disorders.
In patients with positive CES results altered medical management was documented for 13/21 cases highlighting the clinical utility of the assay. Many of these patients in the neurodevelopment category would have received appropriate treatment modalities regardless of CES results. In our experience of this cohort, CES helped to bring an end to the diagnostic odyssey for these patients, directed sub-specialty follow-up and/or appropriate medical rehabilitation.
In conclusion, our results from 51 complex pediatric probands demonstrate the clinical utility of CES which can be a cost-effective diagnostic test in a tertiary pediatric setting. A positive diagnosis could help the physicians in taking medical management decisions and put an end to the diagnostic odyssey of the patients and reduce the burden of expensive multiple tests. For many negative cases, periodic exome reanalysis can provide clarity for an uncertain diagnosis. We anticipate that our findings will encourage research in the direction of incorporation of CES as a first-line diagnostic test for unexplained/heterogeneous genetic disorders encountered in the Emirati population.
Author contributions
AAT was responsible for conception and design of the study; NAM and AI compiled the data; PK and AAT analyzed the data. PK and AAT drafted the manuscript. AAT edited the manuscript. SR, ME, YQ, DH, AT, ZA, MU, AA, WA, and BF provided overall feedback and edited the manuscript.
Funding
No specific funding was available for this study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.04.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013 doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA – J Am Med Assoc. 2014 doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tadmouri GO. Genetic Disorders in the Arab World. In: Tadmouri G, Taleb Al Ali M, Al Khaja N, editors. Genet. Disord. Arab World Qatar, Dubai, United Arab Emirates: Centre for Arab Genomic Studies; 2012.
- 5.Christianson A, Howson C, Modell B. March of Dimes. Global report on birth defect. The hidden toll of dying and disabled children. New York 2006.
- 6.Al-Gazali L., Ali B.R. Mutations of a country: a mutation review of single gene disorders in the United Arab Emirates (UAE) Hum Mutat. 2010;31:505–520. doi: 10.1002/humu.21232. [DOI] [PubMed] [Google Scholar]
- 7.Al-Gazali L., Hamamy H., Al-Arrayad S. Genetic disorders in the Arab world. Br Med J. 2006;333:831–834. doi: 10.1136/bmj.38982.704931.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Shamsi A., Hertecant J.L., Souid A.K., Al-Jasmi F.A. Whole exome sequencing diagnosis of inborn errors of metabolism and other disorders in United Arab Emirates. Orphanet J Rare Dis. 2016;11 doi: 10.1186/s13023-016-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H.C., Tan Q., Rousseaux M.W.C., Wang W., Kim J.Y., Richman R. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49:527–536. doi: 10.1038/ng.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias C., Estruch S.B., Graham S.A., McRae J., Sawiak S.J., Hurst J.A. BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet. 2016;99:253–274. doi: 10.1016/j.ajhg.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajcan-Separovic E., Harvard C., Liu X., McGillivray B., Hall J.G., Qiao Y. Clinical and molecular cytogenetic characterisation of a newly recognised microdeletion syndrome involving 2p15-16.1. J Med Genet. 2007;44:269–276. doi: 10.1136/jmg.2006.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Codipilly D.C., Gavrilova R.H., Tangalos EG. De novo 2p16.1 microdeletion with metastatic esophageal adenocarcinoma. BMJ Case Rep. 2017;2017 doi: 10.1136/bcr-2016-218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abou Tayoun A.N., Krock B., Spinner N.B. Sequencing-based diagnostics for pediatric genetic diseases: progress and potential. Expert Rev Mol Diagn. 2016;16:987–999. doi: 10.1080/14737159.2016.1209411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F. Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA – J Am Med Assoc. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21:2413–2421. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor J.L., Debost J.C.P.G., Morton S.U., Wigdor E.M., Heyne H.O., Lal D. Paternal-age-related de novo mutations and risk for five disorders. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-11039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A. Characterization of greater middle eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–1079. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Gazali L., Hamamy H. Consanguinity and dysmorphology in Arabs. Hum Hered. 2014;77:93–107. doi: 10.1159/000360421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.