Figure 3. Computational approaches to identify snoRNAs.
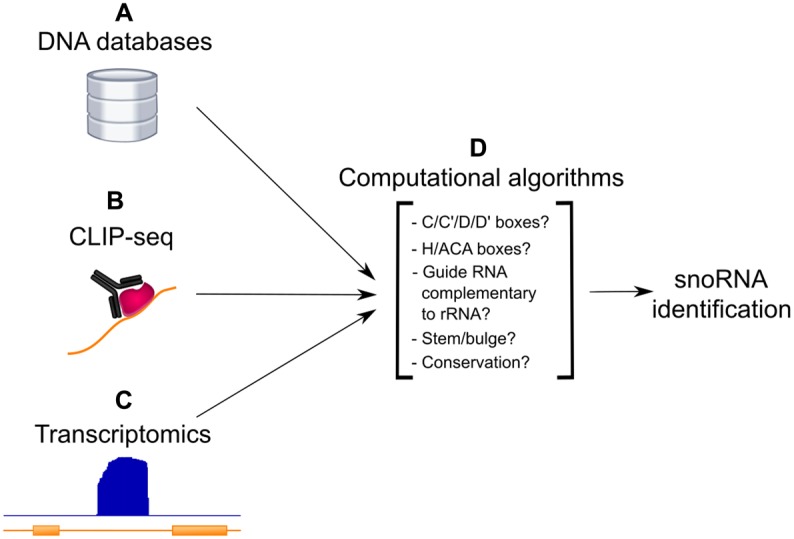
Starting from either sequence databases (A), the transcripts bound to specific proteins as detected by CLIP-seq experiments (B) or non-annotated genomic regions displaying strong levels of expression in RNA-seq datasets (C), computational algorithms consider several different features including, but not limited to, the presence of sequence motifs, complementarity to rRNA, secondary structure features and conservation to predict snoRNAs (D).
