Figure 4. Overview of non-canonical mechanisms of action described for snoRNAs.
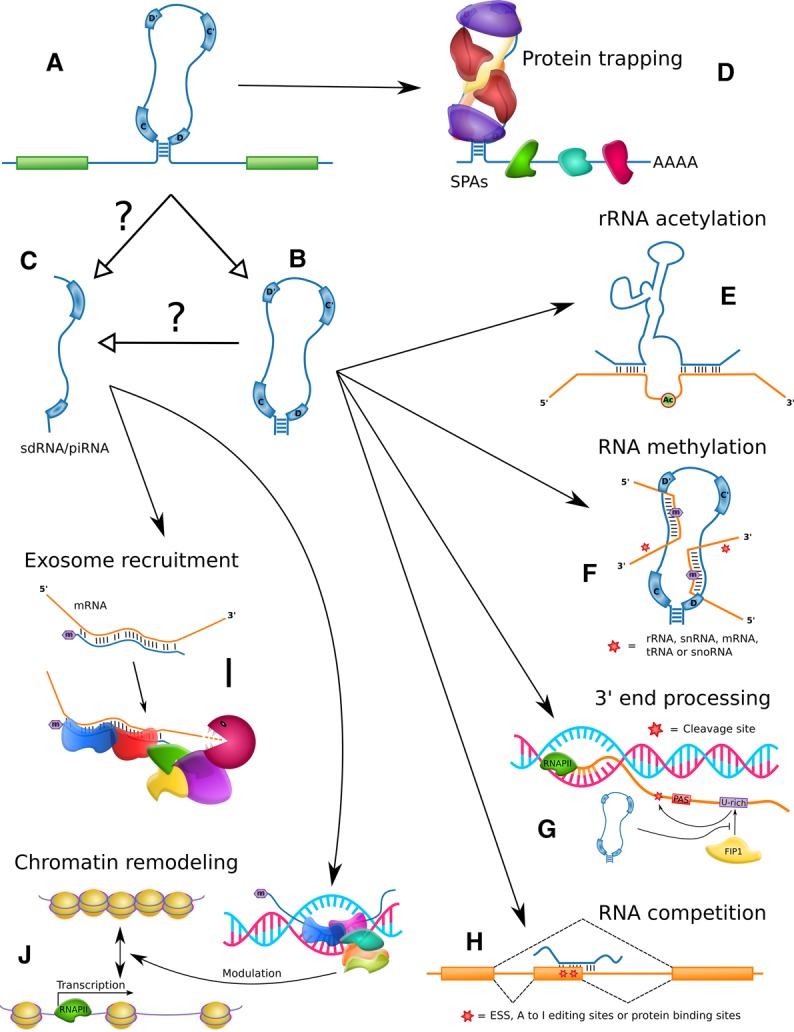
(A) Mammalian snoRNAs are typically embedded in an intron of another gene. (B) Following splicing, intron debranching, protein binding and exonucleolytic degradation, the mature snoRNA is formed. (C) Stable fragments of snoRNAs referred to as sdRNAs for snoRNA-derived RNAs have been detected and could be processed from the mature snoRNA or its precursors. Some sdRNAs have been characterized as piRNAs. (D) Longer noncoding transcripts containing snoRNAs have been found to sequester specific proteins. (E) Some snoRNAs can acetylate rRNA. (F) SnoRNAs can methylate diverse non-canonical substrates including tRNA and mRNA. (G) Specific snoRNAs can bind 3′ end processing protein factors, affecting the choice of polyadenylation sites. (H) SnoRNAs can interact with other RNA, competing for functional binding sites. (I) SdRNAs can regulate pre-mRNA stability through direct binding and recruitment of the nuclear exosome. (J) SdRNAs can also recruit chromatin-modifying complexes to promoters by direct binding. Throughout the figure, white arrowheads indicate processing relationships whereas black arrowheads depict regulatory relationships.
