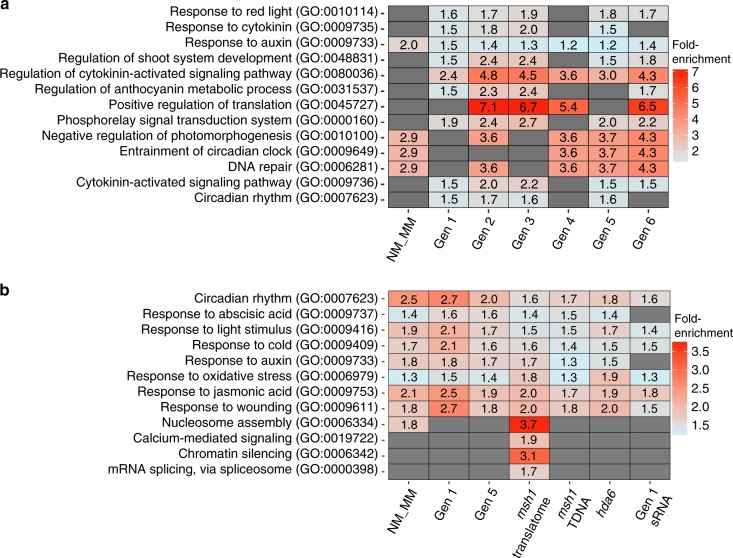
Fig. 5. Statistically significant overenrichment of gene networks.
a Networks identified by NEAT (Network Enrichment Analysis Test) for the DMGs identified in msh1 memory line by MethylIT. The full list of DMGs for each generation of the memory line is listed in Supplementary Data 2. b Functional enrichment analysis of DEGs in msh1 memory line, msh1 mutant translatome and msh1 T-DNA mutant. GO biological process enrichment categories above the cutoff, FDR < 0.01, are shown. DAVID GO was used to conduct the analysis76. The msh1 T-DNA mutant expression data were generated and reported in Shao et al.16. The msh1 mutant translatome refers to the MSH1 cell type-specific TRAPseq dataset that was reported in Beltrán et al.20. The hda6 transcriptome data were taken from Yu et al.27 and reanalyzed using our pipeline. For Gen1 sRNA, genes within ±1 kb of differentially expressed sRNA clusters were used for the GO analysis. Source data for generational DMGs are provided as a Source Data file.