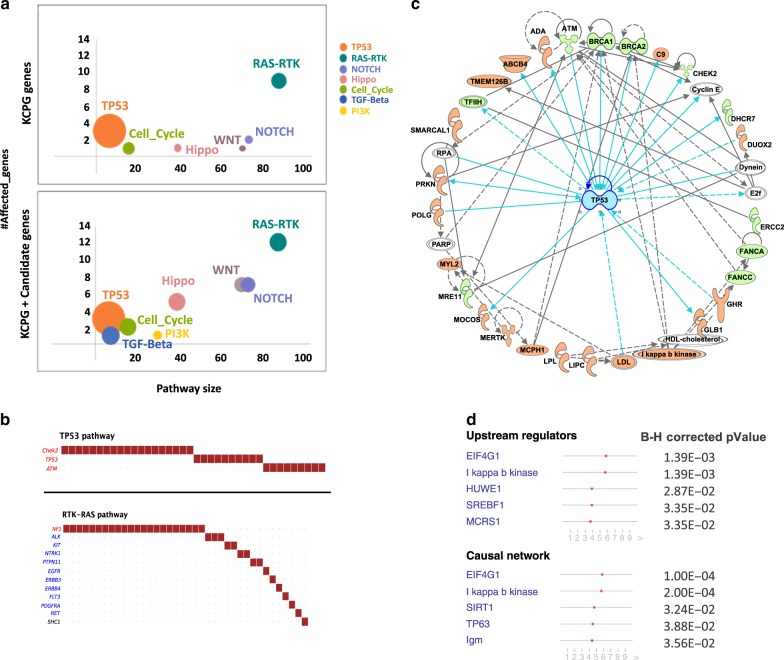
Fig. 4. Pathway analysis of altered genes with germline pathogenic/likely pathogenic (P/LP) variants in children, adolescents, and young adults (C-AYA) with solid tumors.
a Affected pathways based on altered genes with P/LP germline variants. Top panel: only known cancer-predisposing genes (KCPG), lower panel: a combination of all KCPGs and candidate genes. Size of the circles increases as the fraction affected increases. b Genes mutated in TP53 (top panel) and RAS–RTK (lower panel) pathways, and the number of patients affected in our cohort. Red font: tumor suppressor genes; blue font: oncogenes. c Top network, predicted by Ingenuity Pathway Analysis (IPA), based on all the KCPG (green color) and candidate genes (salmon color) with at least four P/LP variants in our C-AYA patients with solid tumors (right-tailed Fisher’s exact test P = 1 × 10−42). d Eukaryotic Translation Initiation Factor 4 Gamma 1 (EIF4G1, B–H corrected P = 1.39 × 10−3) and I kappa b kinase (IκB kinase, B–H corrected P = 1.39 × 10−3) predicted to be the top upstream regulators/causal network based on our IPA analysis. Right-tailed Fisher’s exact test was used, and Benjamini–Hochberg (B–H) P value correction performed to reduce the false discovery rate (FDR).