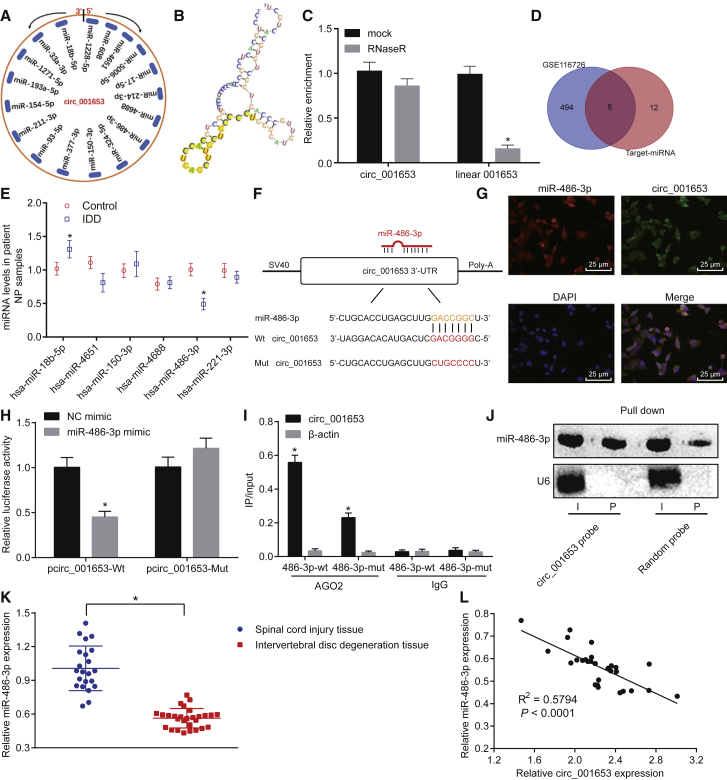
Figure 4.
circ_001653 Regulates miR-486-3p in NPCs
Prediction of miRNAs with binding sites to circ_001653 through the bioinformatics programs (A). Sequence fragments of circ_001653 binding to miR-486-3p (B). The abundance of circ_001653 and linear 001653, with or without treatment of RNase R enzyme (C). Results of the top 500 differentially expressed miRNAs in microarray GSE116726 and miRNAs predicted by bioinformatics. Intersecting miRNAs were hsa-miR-18b-5p, hsa-miR-4651, hsa-miR-150-3p, hsa-miR-4688, hsa-miR-486-3p, and hsa-miR-221-3p (D). Relative expression of miRNAs detected by qRT-PCR. (E). The binding sites of circ_001653 to miR-486-3p analyzed by the bioinformatics programs (F). circ_001653 and miR-486-3p were located in the cytoplasm of NPCs, as detected by RNA FISH; scale bars, 25 μm (G). Identification of interaction between circ_001653 and miR-486-3p by the Dual Luciferase Reporter Gene Assay (H). AGO2 immunoprecipitation in degenerative NPCs transfected with miR-486-3p mimic or NC and the quantification of miR-486-3p expression determined by qRT-PCR (I). Northern blot analysis of miR-486-3p after being pulled down by a circ_001653 probe or a random probe (J). The relative expression of miR-486-3p in NP tissues of IDD and SCI tissues evaluated by qRT-PCR (K). The correlation between circ_001653 and miR-486-3p assessed by Pearson’s correlation analysis (L). *p < 0.05 versus NC mimic or SCI tissues. Statistical data in Figures 5J and 5K were measurement data described as mean ± standard deviation; n = 30 for the SCI tissues, and n = 36 for the NP tissues from the IDD patients. The experiment was repeated three times independently. Data between two groups were analyzed by t test. The one-way ANOVA was adopted for comparison among multiple groups, followed by Tukey’s post hoc test.