Figure 1.
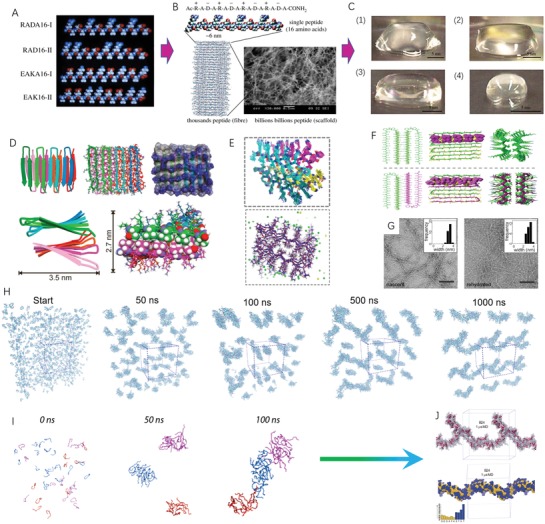
Designer self‐assembling peptide systems and molecular design strategy. A) The molecular models of partial designer peptide moduli. Single molecular building block harbors one hydrophobic side and another hydrophilic side in each modulus, respectively. B) RADA16‐I peptide is at the dimensions with ≈6 nm long, 1.3 nm wide, 0.8 nm thick, and with the charge arrangement ± by four times repeat on the hydrophilic side. Approximately hundreds of thousands or millions of RADA16‐I peptides self‐assemble into a nanofiber architecture depending on the fiber length as revealed by the SEM image. C) Schematic images of RADA16‐I peptide hydrogel at various conditions: (1) 0.5 wt% (pH 7.5), (2) 0.1 wt% (pH 7.5, Tris–HCl), (3) 0.1 wt% (pH 7.5, PBS) before sonication and (4) reassembled RADA16‐I peptide hydrogel after four times of sonication. Reproduced with permission.[ 20 ] Copyright 2017, The Royal Chemical Society. D) Cartoon, tube, and stick models indicate the molecular relationship between β hairpins in the MAX1 peptide fibrils. E) Superposition of MAX1 peptide structures from 20 equally spaced frames from the final half of the molecular dynamics/Monte Carlo trajectory (upper panel) and final MAX1 fibril structure from molecular dynamics/Monte Carlo calculations (low panel).[ 28a ] F) A proposed molecular dynamic model of MAX1/DMAX1 in their coassembled, racemic fibrillar state. ChemDraw figures at top define strand orientation in each assembly. Central images highlight the relative orientation of hairpins within a single monolayer of each fibril type with the valine side chains rendered in CPK (magenta). Bottom images view the racemic and pure enantiomeric fibrils along their long axes.[ 28b ] G) Characterization of MAX1 fibrils by TEM and solid‐state NMR. Negatively stained TEM images of nascent MAX1 fibrils (left) and fibrils in a rehydrated hydrogel after lyophilization (right). (Insets) Average nanofibril widths for each sample were ≈3.5 nm (Scale bar = 100 nm). Panels (D–G) are reproduced with permission.[ 28 ] Copyright 2017, American Chemical Society; Copyright 2015, National Academy of Sciences, USA. H) All snapshots are obtained from a 1 µs restrained atomistic MD simulation of 100 B24 peptides that are initially randomly placed in a water‐filled periodic box. I) Snapshots of B24 peptides, illustrating the preference to assemble into smaller elongated rod‐like clusters that then assemble to larger fibers. J) The supramolecular organization, B24 fiber‐like assembly after 1 µs of restrained atomic MD simulation. Panels (G–I) reproduced with permission.[ 29 ] Copyright 2019, Wiley‐VCH.
